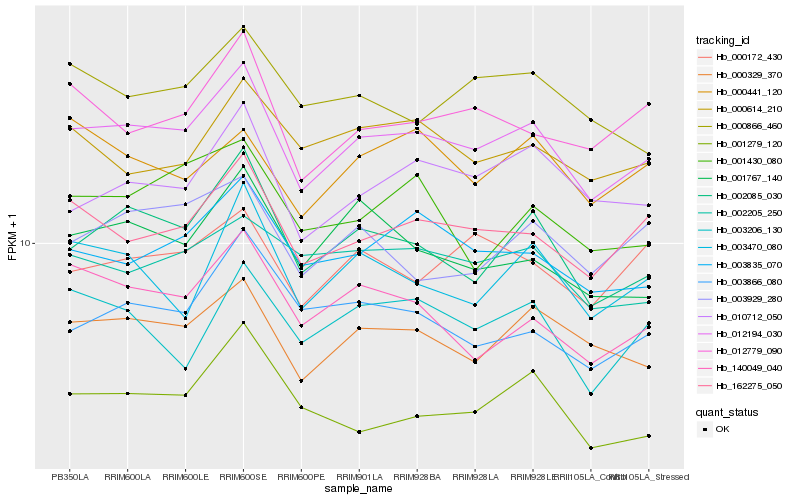
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012194_030 |
0.0 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000329_370 |
0.0472630199 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 3 |
Hb_162275_050 |
0.0475172942 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 4 |
Hb_002085_030 |
0.0485850195 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003835_070 |
0.0544274597 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010712_050 |
0.0570672405 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 7 |
Hb_012779_090 |
0.0601586213 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 8 |
Hb_002205_250 |
0.0618543925 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001279_120 |
0.0620223048 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 10 |
Hb_001767_140 |
0.0622536725 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 11 |
Hb_140049_040 |
0.0624907046 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 12 |
Hb_000172_430 |
0.0634459646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003470_080 |
0.0638200517 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000614_210 |
0.0651278813 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003866_080 |
0.0652389097 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 16 |
Hb_003929_280 |
0.0653383078 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 17 |
Hb_003206_130 |
0.0654254912 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 18 |
Hb_000866_460 |
0.0657129008 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001430_080 |
0.0660247763 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 20 |
Hb_000441_120 |
0.0660986139 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |