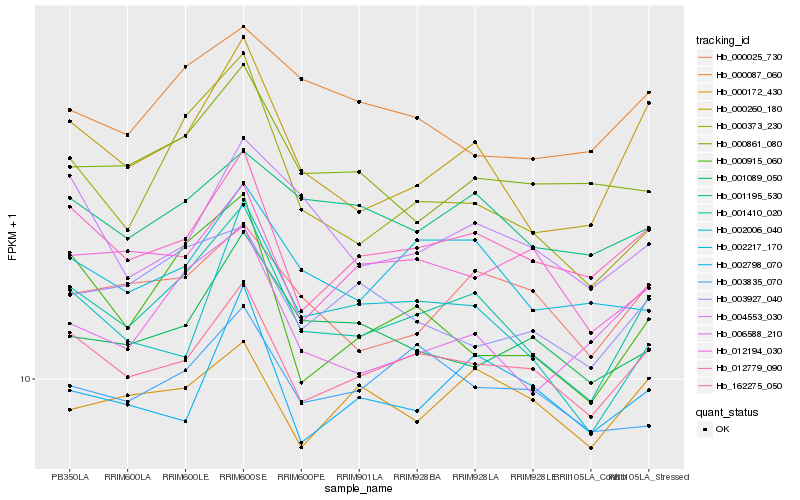
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001410_020 |
0.0 |
- |
- |
cdk8, putative [Ricinus communis] |
| 2 |
Hb_000260_180 |
0.0518931222 |
- |
- |
PREDICTED: KH domain-containing protein At2g38610 [Jatropha curcas] |
| 3 |
Hb_162275_050 |
0.0565147601 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 4 |
Hb_012779_090 |
0.0623189519 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 5 |
Hb_000373_230 |
0.0672472042 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 6 |
Hb_006588_210 |
0.0694534882 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 7 |
Hb_000025_730 |
0.0710915454 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002217_170 |
0.073226028 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 9 |
Hb_001195_530 |
0.0733602742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000915_060 |
0.0735709371 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 11 |
Hb_003927_040 |
0.076528929 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 12 |
Hb_001089_050 |
0.0785241115 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_002006_040 |
0.0793672771 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 14 |
Hb_004553_030 |
0.0799093309 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 15 |
Hb_000087_060 |
0.0799567295 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_000861_080 |
0.0801421117 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 17 |
Hb_003835_070 |
0.0802305546 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012194_030 |
0.080504053 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002798_070 |
0.0818158561 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 20 |
Hb_000172_430 |
0.0821921279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |