| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
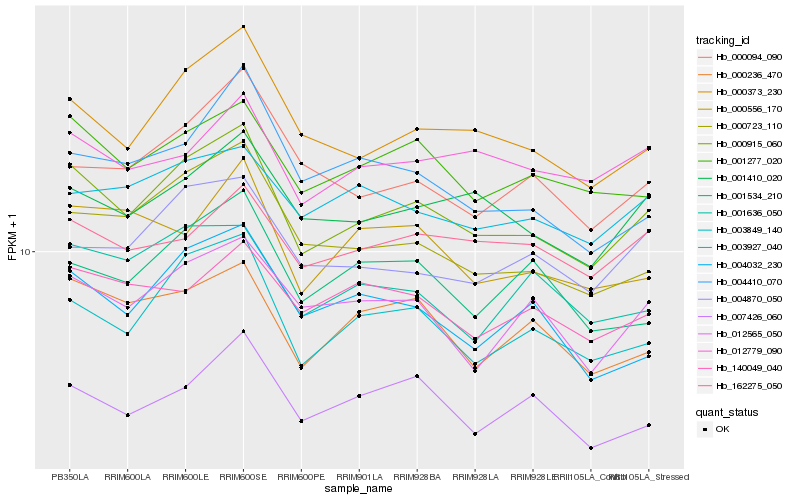
Hb_000915_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 2 |
Hb_000373_230 |
0.062296236 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 3 |
Hb_003849_140 |
0.0624610442 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 4 |
Hb_001277_020 |
0.0694691564 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 5 |
Hb_000723_110 |
0.0728001524 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 6 |
Hb_001410_020 |
0.0735709371 |
- |
- |
cdk8, putative [Ricinus communis] |
| 7 |
Hb_000236_470 |
0.0797419927 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 8 |
Hb_012565_050 |
0.0798110484 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 9 |
Hb_004870_050 |
0.0813371503 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 10 |
Hb_004032_230 |
0.0852094962 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 11 |
Hb_003927_040 |
0.0855669057 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 12 |
Hb_001636_050 |
0.0857788438 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 13 |
Hb_000094_090 |
0.0870892764 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007426_060 |
0.0871833295 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_001534_210 |
0.0872020553 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000556_170 |
0.0886949155 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 17 |
Hb_140049_040 |
0.0888734186 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 18 |
Hb_004410_070 |
0.0888973038 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 19 |
Hb_012779_090 |
0.0889396117 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 20 |
Hb_162275_050 |
0.0890280651 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |