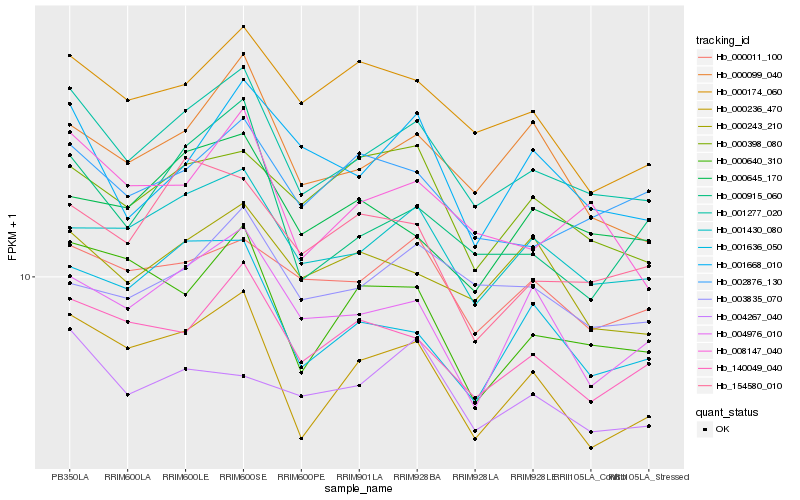
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_020 |
0.0 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 2 |
Hb_001430_080 |
0.0545997373 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 3 |
Hb_000236_470 |
0.056736097 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 4 |
Hb_140049_040 |
0.0593794042 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 5 |
Hb_008147_040 |
0.0593795627 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 6 |
Hb_000099_040 |
0.0644932476 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000011_100 |
0.0660927169 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_002876_130 |
0.0669271642 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 9 |
Hb_000174_060 |
0.068092363 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 10 |
Hb_000915_060 |
0.0694691564 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 11 |
Hb_004976_010 |
0.0699727329 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 12 |
Hb_000243_210 |
0.0710829453 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000398_080 |
0.0718355639 |
- |
- |
tip120, putative [Ricinus communis] |
| 14 |
Hb_000640_310 |
0.0727531094 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 15 |
Hb_003835_070 |
0.0742298346 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 16 |
Hb_154580_010 |
0.074780741 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 17 |
Hb_001636_050 |
0.0749410033 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 18 |
Hb_000645_170 |
0.0749755892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001668_010 |
0.075113812 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 20 |
Hb_004267_040 |
0.0752848358 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |