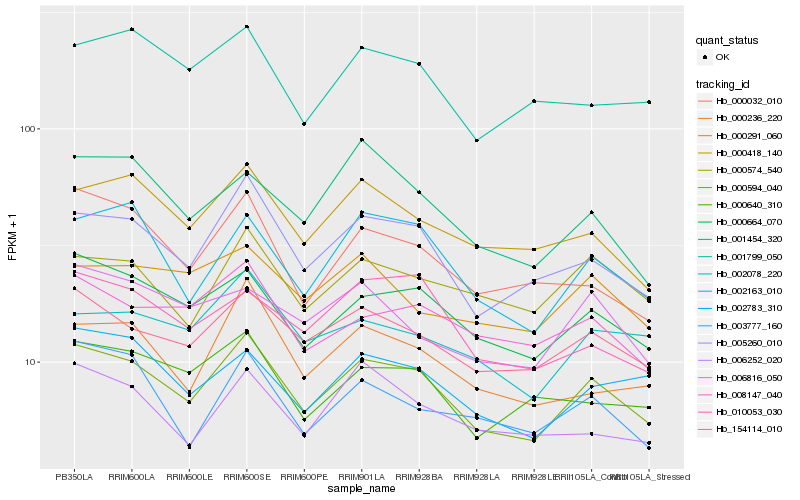
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_040 |
0.0 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 2 |
Hb_005260_010 |
0.0554159284 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_154114_010 |
0.0569920016 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 4 |
Hb_000664_070 |
0.0572836894 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 5 |
Hb_002163_010 |
0.0702371129 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 6 |
Hb_000418_140 |
0.0735446858 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 7 |
Hb_008147_040 |
0.0779904302 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 8 |
Hb_002783_310 |
0.0785839774 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 9 |
Hb_000640_310 |
0.0788052515 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 10 |
Hb_000574_540 |
0.0789151241 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000236_220 |
0.0815216025 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 12 |
Hb_010053_030 |
0.0817531283 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 13 |
Hb_002078_220 |
0.081865685 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006252_020 |
0.0825643495 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000032_010 |
0.0830097061 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 16 |
Hb_001799_050 |
0.0831183506 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 17 |
Hb_003777_160 |
0.0843902163 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006816_050 |
0.0849371502 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_000291_060 |
0.0856669055 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_001454_320 |
0.0858567937 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |