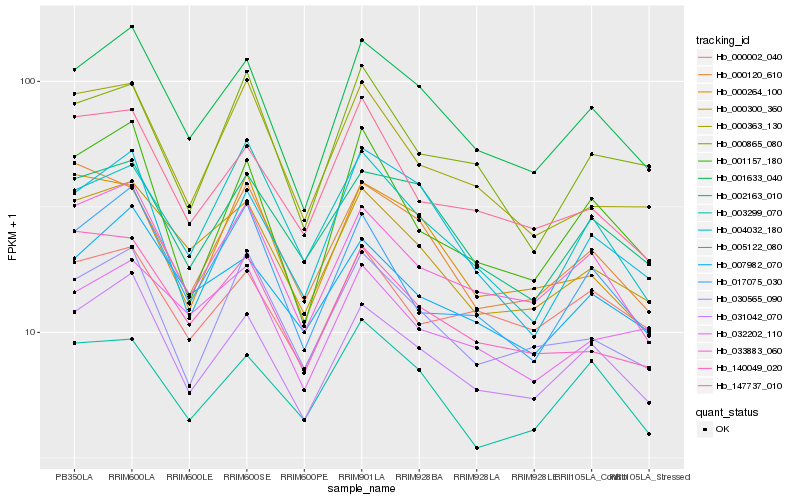
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001633_040 |
0.0 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 2 |
Hb_031042_070 |
0.0430773278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_033883_060 |
0.060857559 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 4 |
Hb_000300_360 |
0.0622561901 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 5 |
Hb_007982_070 |
0.0723263857 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 6 |
Hb_002163_010 |
0.0728597509 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 7 |
Hb_030565_090 |
0.0779293828 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 8 |
Hb_005122_080 |
0.0816642185 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 9 |
Hb_017075_030 |
0.0846318998 |
- |
- |
helicase, putative [Ricinus communis] |
| 10 |
Hb_001157_180 |
0.0854444256 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_003299_070 |
0.0859232989 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 12 |
Hb_032202_110 |
0.088203279 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 13 |
Hb_140049_020 |
0.0885333542 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 14 |
Hb_147737_010 |
0.0898043151 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000120_610 |
0.0908266656 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 16 |
Hb_000002_040 |
0.0908327727 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 17 |
Hb_004032_180 |
0.0919106951 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 18 |
Hb_000865_080 |
0.0924645641 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 19 |
Hb_000264_100 |
0.0925037644 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000363_130 |
0.0929759079 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |