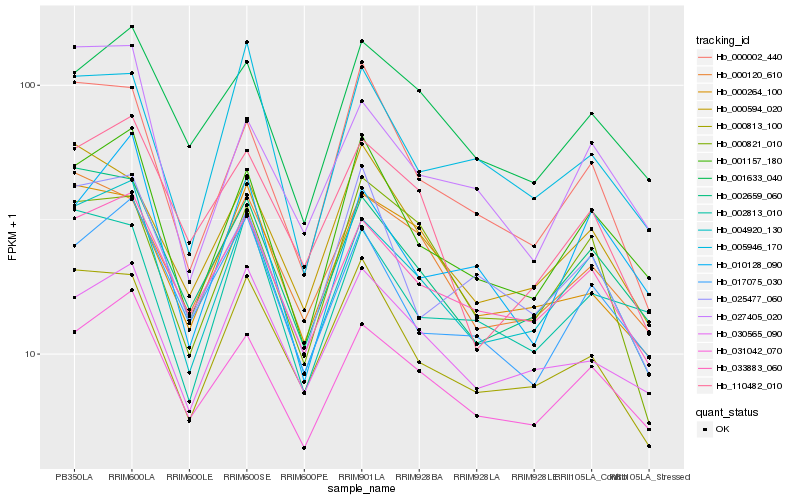
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004920_130 |
0.0 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002659_060 |
0.0669413133 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 3 |
Hb_033883_060 |
0.0675751388 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 4 |
Hb_001157_180 |
0.0720740386 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_031042_070 |
0.0748115457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000813_100 |
0.0855891121 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000821_010 |
0.0898006507 |
- |
- |
PREDICTED: auxin response factor 19-like [Jatropha curcas] |
| 8 |
Hb_000120_610 |
0.089821913 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 9 |
Hb_030565_090 |
0.0916921363 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 10 |
Hb_000264_100 |
0.0930786679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_017075_030 |
0.093195858 |
- |
- |
helicase, putative [Ricinus communis] |
| 12 |
Hb_000594_020 |
0.093312836 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001633_040 |
0.0933943181 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 14 |
Hb_000002_440 |
0.0935988632 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 15 |
Hb_110482_010 |
0.0938814215 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 16 |
Hb_027405_020 |
0.0947628826 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 17 |
Hb_010128_090 |
0.0956705338 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 18 |
Hb_005946_170 |
0.0999206859 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 19 |
Hb_002813_010 |
0.1006460561 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 20 |
Hb_025477_060 |
0.1032072568 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |