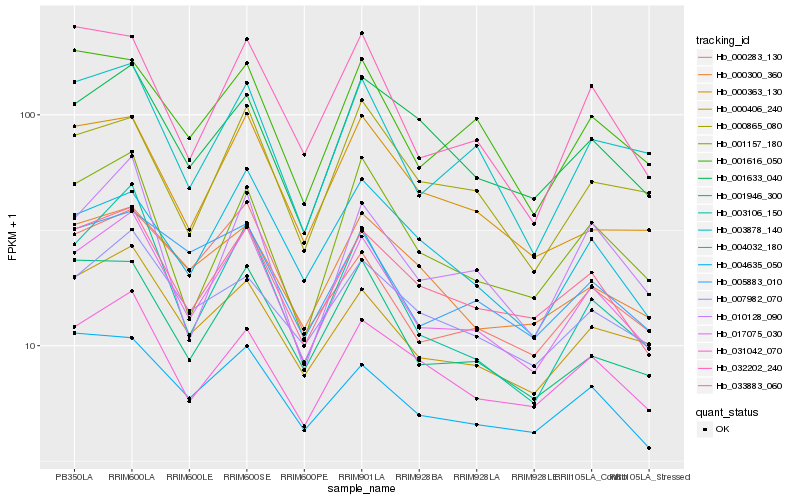
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017075_030 |
0.0 |
- |
- |
helicase, putative [Ricinus communis] |
| 2 |
Hb_033883_060 |
0.0658357682 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 3 |
Hb_000406_240 |
0.0689832488 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 4 |
Hb_003106_150 |
0.0728752863 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 5 |
Hb_031042_070 |
0.0770955267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001946_300 |
0.0777537139 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 7 |
Hb_003878_140 |
0.0795889246 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 8 |
Hb_000283_130 |
0.0796543408 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 9 |
Hb_001616_050 |
0.0822773728 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 10 |
Hb_005883_010 |
0.083696927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001157_180 |
0.0838311221 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_032202_240 |
0.084238596 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 13 |
Hb_001633_040 |
0.0846318998 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 14 |
Hb_004032_180 |
0.0849309975 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 15 |
Hb_000363_130 |
0.0850140144 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 16 |
Hb_007982_070 |
0.0855026713 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 17 |
Hb_010128_090 |
0.0877798365 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 18 |
Hb_000300_360 |
0.087932687 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 19 |
Hb_000865_080 |
0.0883282231 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 20 |
Hb_004635_050 |
0.0887979776 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |