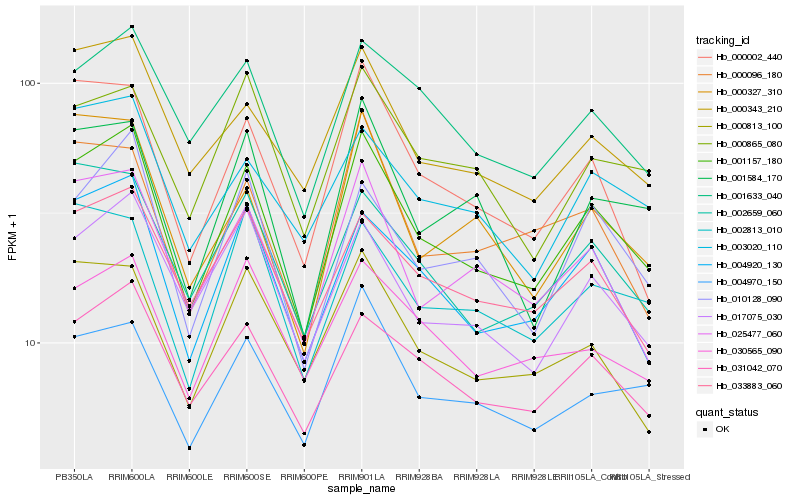
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_180 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_004920_130 |
0.0720740386 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_010128_090 |
0.0782941748 |
- |
- |
PREDICTED: uncharacterized protein LOC105647377 [Jatropha curcas] |
| 4 |
Hb_031042_070 |
0.0794072387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000343_210 |
0.0835533849 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 6 |
Hb_017075_030 |
0.0838311221 |
- |
- |
helicase, putative [Ricinus communis] |
| 7 |
Hb_000002_440 |
0.0843991361 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 8 |
Hb_025477_060 |
0.0844153519 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 9 |
Hb_001633_040 |
0.0854444256 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 10 |
Hb_002659_060 |
0.08599024 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 11 |
Hb_033883_060 |
0.0870022713 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 12 |
Hb_004970_150 |
0.0893079883 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 13 |
Hb_002813_010 |
0.090125929 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000865_080 |
0.0906106475 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 15 |
Hb_001584_170 |
0.0915315192 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 16 |
Hb_003020_110 |
0.0929751572 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_000813_100 |
0.0940087681 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 18 |
Hb_030565_090 |
0.0941148495 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 19 |
Hb_000327_310 |
0.0943319834 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 20 |
Hb_000096_180 |
0.0946904357 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g67570, chloroplastic-like isoform X1 [Jatropha curcas] |