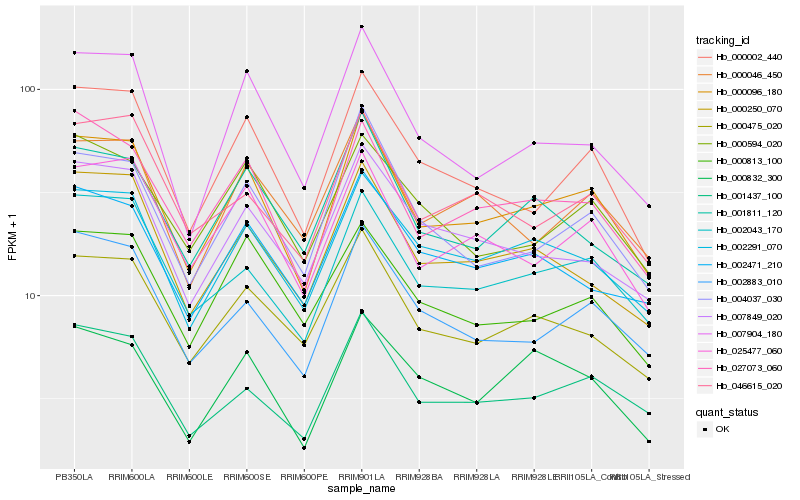
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000096_180 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g67570, chloroplastic-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_027073_060 |
0.0688223186 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_025477_060 |
0.0703610982 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 4 |
Hb_002291_070 |
0.0708336651 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Prunus mume] |
| 5 |
Hb_002043_170 |
0.0711663651 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 6 |
Hb_001437_100 |
0.0720120823 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 7 |
Hb_000002_440 |
0.0720958721 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 8 |
Hb_000475_020 |
0.0772781326 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_004037_030 |
0.0794903825 |
- |
- |
PREDICTED: DNA repair endonuclease UVH1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002883_010 |
0.0797600943 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000046_450 |
0.0811554196 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 12 |
Hb_007904_180 |
0.0815462829 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Jatropha curcas] |
| 13 |
Hb_000250_070 |
0.0824142828 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase listerin [Jatropha curcas] |
| 14 |
Hb_007849_020 |
0.0869398392 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 15 |
Hb_001811_120 |
0.0872238252 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002471_210 |
0.0874585745 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 17 |
Hb_000832_300 |
0.0880236468 |
- |
- |
- |
| 18 |
Hb_046615_020 |
0.0903098692 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 19 |
Hb_000813_100 |
0.0912918395 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000594_020 |
0.0926794332 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |