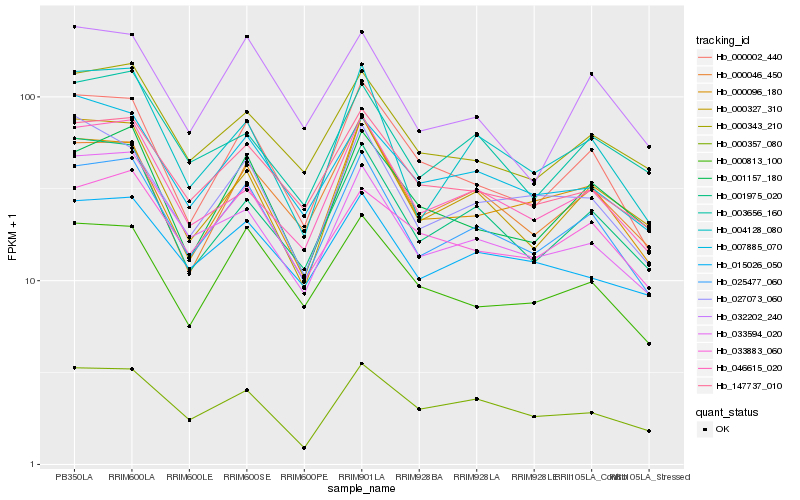
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025477_060 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 2 |
Hb_000002_440 |
0.0702521012 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 3 |
Hb_147737_010 |
0.0702632413 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000096_180 |
0.0703610982 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g67570, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000046_450 |
0.0735222297 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 6 |
Hb_046615_020 |
0.0743017482 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 7 |
Hb_001975_020 |
0.0764729581 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 8 |
Hb_000813_100 |
0.076947039 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004128_080 |
0.0775953661 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 10 |
Hb_000343_210 |
0.0785363804 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 11 |
Hb_033594_020 |
0.080332529 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 12 |
Hb_007885_070 |
0.0819632434 |
- |
- |
cleavage and polyadenylation specificity factor, putative [Ricinus communis] |
| 13 |
Hb_000327_310 |
0.0823675419 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 14 |
Hb_027073_060 |
0.0825594646 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_015026_050 |
0.0834808337 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000357_080 |
0.0842159966 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59200, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001157_180 |
0.0844153519 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_033883_060 |
0.0859018261 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 19 |
Hb_003656_160 |
0.0887012802 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein B456_013G226400 [Gossypium raimondii] |
| 20 |
Hb_032202_240 |
0.0889584393 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |