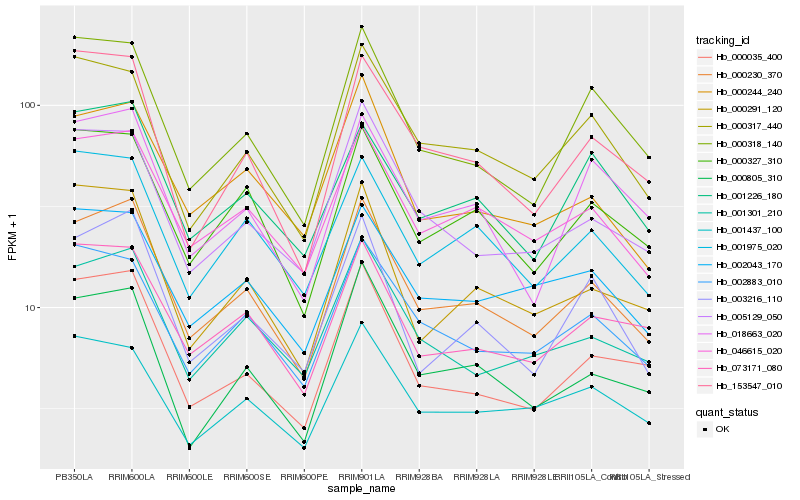
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_370 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_046615_020 |
0.0604295081 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 3 |
Hb_000327_310 |
0.0689675859 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 4 |
Hb_001301_210 |
0.0745896138 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 32 [Jatropha curcas] |
| 5 |
Hb_002883_010 |
0.0783921924 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001975_020 |
0.0791944203 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 7 |
Hb_000318_140 |
0.0793386298 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_003216_110 |
0.0795461276 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 9 |
Hb_000317_440 |
0.0797138332 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000244_240 |
0.0801747471 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_073171_080 |
0.0803537219 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_005129_050 |
0.0812291908 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase [Jatropha curcas] |
| 13 |
Hb_153547_010 |
0.0812872938 |
- |
- |
PREDICTED: protein FLX-like 4 [Jatropha curcas] |
| 14 |
Hb_000035_400 |
0.0817940848 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Citrus sinensis] |
| 15 |
Hb_002043_170 |
0.0820136397 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 16 |
Hb_000805_310 |
0.0835979149 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000291_120 |
0.0853886204 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 18 |
Hb_001437_100 |
0.0858158982 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 19 |
Hb_018663_020 |
0.0862090923 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001226_180 |
0.086861298 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |