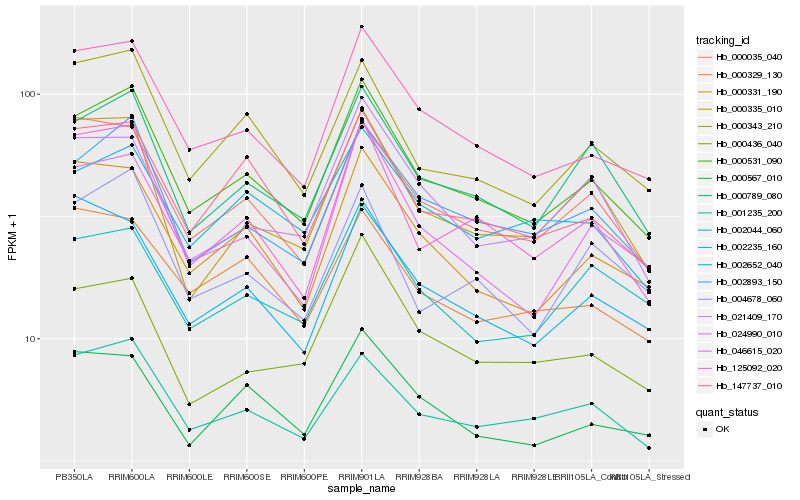
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000531_090 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 2 |
Hb_125092_020 |
0.0457605275 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002893_150 |
0.04643538 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 4 |
Hb_000789_080 |
0.0531625728 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000035_040 |
0.059906738 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 6 |
Hb_000343_210 |
0.0617741579 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 7 |
Hb_147737_010 |
0.0625941229 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000331_190 |
0.0636336994 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 9 |
Hb_024990_010 |
0.0653436592 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 10 |
Hb_046615_020 |
0.0664619337 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 11 |
Hb_001235_200 |
0.0696670337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000567_010 |
0.0711674061 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 13 |
Hb_002235_160 |
0.0748222687 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 14 |
Hb_002652_040 |
0.0753544171 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 15 |
Hb_021409_170 |
0.0772962549 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_004678_060 |
0.0773553278 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 17 |
Hb_000436_040 |
0.0780507536 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 18 |
Hb_000329_130 |
0.0790440414 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 19 |
Hb_000335_010 |
0.0790695827 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_002044_060 |
0.0790937304 |
- |
- |
elongation factor ts, putative [Ricinus communis] |