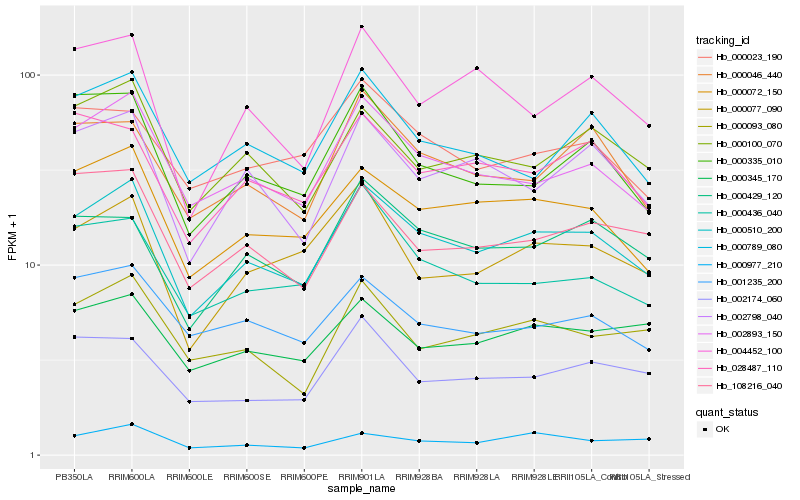
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_200 |
0.0 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 2 |
Hb_000072_150 |
0.0695074676 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 3 |
Hb_002893_150 |
0.0699388889 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 4 |
Hb_002798_040 |
0.0864609239 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 5 |
Hb_000429_120 |
0.0896879662 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 6 |
Hb_000100_070 |
0.0916310923 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ISY1 homolog [Jatropha curcas] |
| 7 |
Hb_000046_440 |
0.092548084 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 8 |
Hb_028487_110 |
0.0945791998 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 9 |
Hb_000789_080 |
0.0949636564 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000077_090 |
0.0953677875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001235_200 |
0.098129299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000093_080 |
0.0994342301 |
- |
- |
hypothetical protein POPTR_0008s09730g [Populus trichocarpa] |
| 13 |
Hb_000023_190 |
0.099665171 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 14 |
Hb_000436_040 |
0.100206952 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 15 |
Hb_004452_100 |
0.1002494322 |
- |
- |
PREDICTED: uncharacterized protein LOC105639563 [Jatropha curcas] |
| 16 |
Hb_108216_040 |
0.1009774006 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 17 |
Hb_000335_010 |
0.1014337825 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_000977_210 |
0.1014665989 |
- |
- |
chromatin assembly factor 1, subunit A, putative [Ricinus communis] |
| 19 |
Hb_000345_170 |
0.1016003567 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002174_060 |
0.1020333984 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |