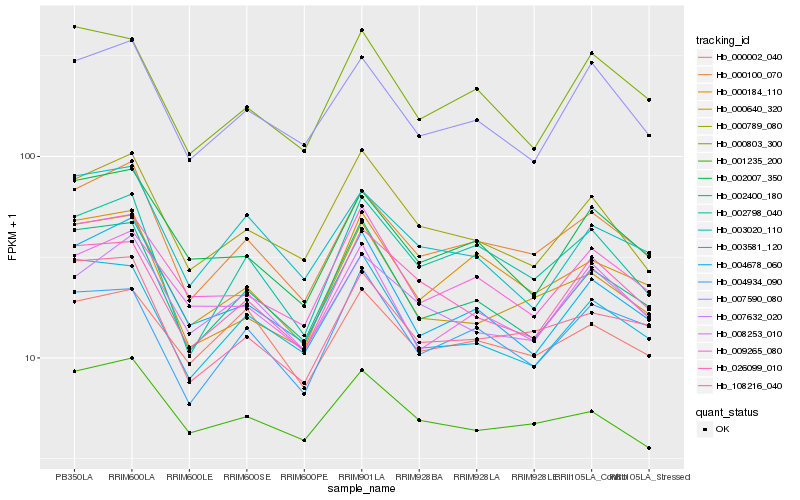
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000100_070 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ISY1 homolog [Jatropha curcas] |
| 2 |
Hb_108216_040 |
0.0549441553 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_002798_040 |
0.0640944096 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 4 |
Hb_007590_080 |
0.0678055631 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-4 [Malus domestica] |
| 5 |
Hb_002400_180 |
0.0710074232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000640_320 |
0.0728397576 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002007_350 |
0.0751393531 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 8 |
Hb_007632_020 |
0.078330099 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 9 |
Hb_001235_200 |
0.0804470887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000184_110 |
0.0808024335 |
- |
- |
hypothetical protein F383_18769 [Gossypium arboreum] |
| 11 |
Hb_004678_060 |
0.0814348318 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 12 |
Hb_009265_080 |
0.0828019997 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_000789_080 |
0.0832248342 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000803_300 |
0.0849839899 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 15 |
Hb_000002_040 |
0.0853668282 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 16 |
Hb_003020_110 |
0.085979767 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_004934_090 |
0.0863418052 |
- |
- |
hypothetical protein JCGZ_08073 [Jatropha curcas] |
| 18 |
Hb_003581_120 |
0.0875993319 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 19 |
Hb_026099_010 |
0.0887725088 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 20 |
Hb_008253_010 |
0.0892812107 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |