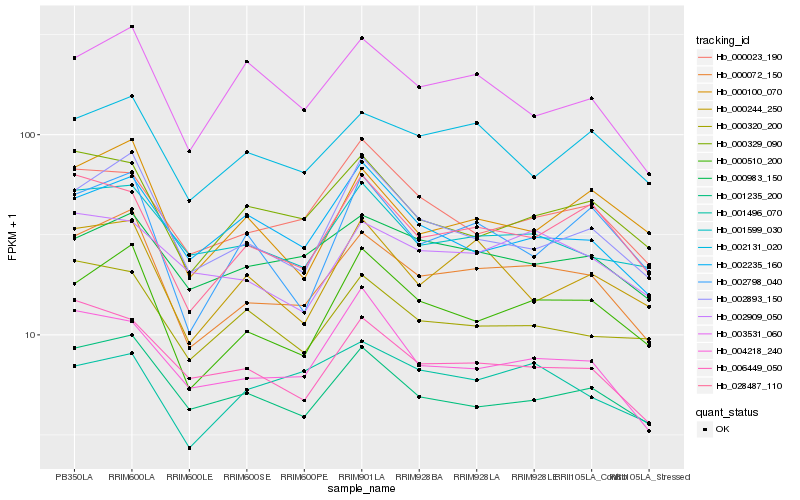
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_150 |
0.0 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 2 |
Hb_000510_200 |
0.0695074676 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 3 |
Hb_002893_150 |
0.0866276778 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 4 |
Hb_001235_200 |
0.0894956128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_028487_110 |
0.0908040224 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 6 |
Hb_001599_030 |
0.0945405892 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_002131_020 |
0.0973050702 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 8 |
Hb_000244_250 |
0.0987360691 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000100_070 |
0.0993980213 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ISY1 homolog [Jatropha curcas] |
| 10 |
Hb_000983_150 |
0.1003352339 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 11 |
Hb_000329_090 |
0.1005774826 |
- |
- |
arginase, putative [Ricinus communis] |
| 12 |
Hb_006449_050 |
0.1013822618 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 13 |
Hb_003531_060 |
0.1015638787 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_002798_040 |
0.1038627582 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 15 |
Hb_004218_240 |
0.1054344445 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 16 |
Hb_000320_200 |
0.1058019433 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 17 |
Hb_002909_050 |
0.1061628756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000023_190 |
0.1074096942 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 19 |
Hb_002235_160 |
0.1099261266 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 20 |
Hb_001496_070 |
0.1102178416 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |