| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
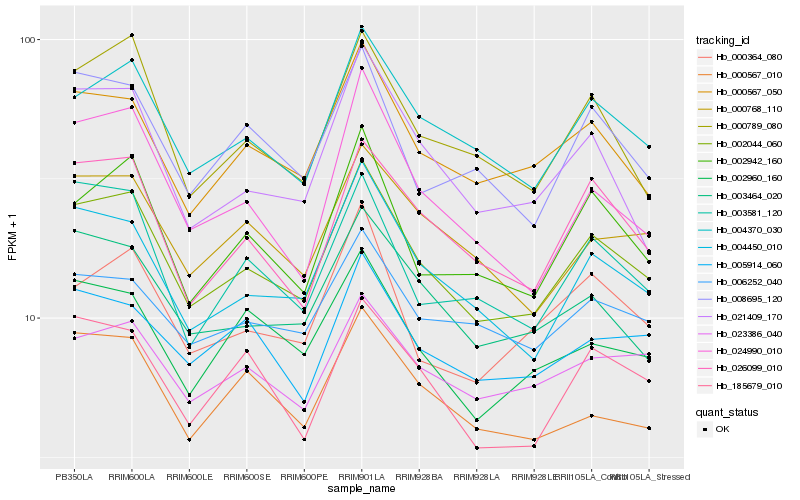
Hb_002044_060 |
0.0 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 2 |
Hb_004370_030 |
0.0530241182 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 3 |
Hb_004450_010 |
0.0554555087 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 4 |
Hb_000567_050 |
0.0606151428 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 5 |
Hb_026099_010 |
0.063722367 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 6 |
Hb_002942_160 |
0.0649429453 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 7 |
Hb_000789_080 |
0.0666211035 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003464_020 |
0.068935866 |
- |
- |
srpk, putative [Ricinus communis] |
| 9 |
Hb_000768_110 |
0.0698841545 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_185679_010 |
0.0712070234 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 11 |
Hb_003581_120 |
0.0715338466 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 12 |
Hb_008695_120 |
0.0737701383 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_002960_160 |
0.0745386499 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 14 |
Hb_023386_040 |
0.0751370622 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 15 |
Hb_000364_080 |
0.0759993764 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_021409_170 |
0.0760051974 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_006252_040 |
0.076155926 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000567_010 |
0.0761722244 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 19 |
Hb_005914_060 |
0.0763517793 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 20 |
Hb_024990_010 |
0.076401646 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |