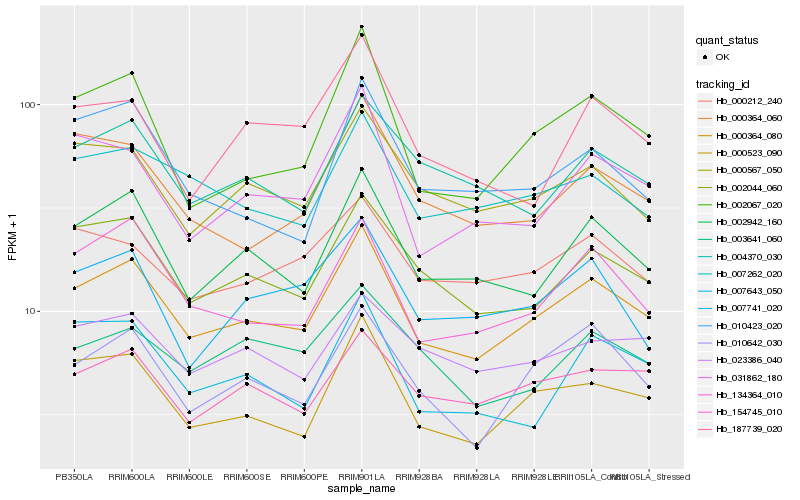
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000364_080 |
0.0 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002942_160 |
0.0687508385 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 3 |
Hb_000523_090 |
0.0715115387 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 4 |
Hb_002044_060 |
0.0759993764 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 5 |
Hb_134364_010 |
0.0790522302 |
- |
- |
PREDICTED: transcriptional adapter ADA2b isoform X1 [Jatropha curcas] |
| 6 |
Hb_002067_020 |
0.0794126375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010642_030 |
0.0827045001 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_000567_050 |
0.085150889 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 9 |
Hb_031862_180 |
0.0876862644 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 10 |
Hb_154745_010 |
0.0884900307 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_007262_020 |
0.0889105466 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 12 |
Hb_004370_030 |
0.093015916 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 13 |
Hb_000364_060 |
0.0937179758 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 14 |
Hb_000212_240 |
0.0964677203 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_023386_040 |
0.0980889542 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 16 |
Hb_007741_020 |
0.0985113835 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 17 |
Hb_007643_050 |
0.0998450802 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_010423_020 |
0.1010497013 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 19 |
Hb_003641_060 |
0.1017605397 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 20 |
Hb_187739_020 |
0.1020782297 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |