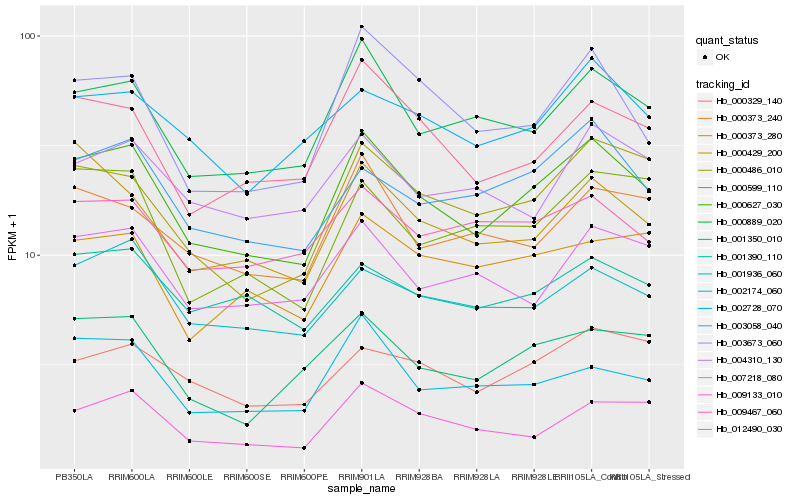
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000627_030 |
0.0 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 2 |
Hb_000889_020 |
0.0789591779 |
- |
- |
ribosome biogenesis protein brix, putative [Ricinus communis] |
| 3 |
Hb_012490_030 |
0.0867005624 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000486_010 |
0.0867347366 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003058_040 |
0.0922769293 |
- |
- |
rotamase, putative [Ricinus communis] |
| 6 |
Hb_003673_060 |
0.0927084641 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 7 |
Hb_009133_010 |
0.0942519884 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase-like [Jatropha curcas] |
| 8 |
Hb_001936_060 |
0.0952650062 |
- |
- |
pyroglutamyl-peptidase I, putative [Ricinus communis] |
| 9 |
Hb_001350_010 |
0.0953011852 |
- |
- |
PREDICTED: uncharacterized protein LOC105629383 [Jatropha curcas] |
| 10 |
Hb_002174_060 |
0.0969638014 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 11 |
Hb_000373_240 |
0.0980288213 |
- |
- |
PREDICTED: uncharacterized protein LOC105640940 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000329_140 |
0.0984039648 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 13 |
Hb_007218_080 |
0.0992413205 |
- |
- |
PREDICTED: uncharacterized protein LOC105631523 [Jatropha curcas] |
| 14 |
Hb_002728_070 |
0.0996145054 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 15 |
Hb_000599_110 |
0.1024822617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000429_200 |
0.1025148551 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 17 |
Hb_000373_280 |
0.104268111 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009467_060 |
0.1045432585 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001390_110 |
0.1052052754 |
- |
- |
PREDICTED: uncharacterized protein LOC105635984 [Jatropha curcas] |
| 20 |
Hb_004310_130 |
0.105741058 |
- |
- |
PREDICTED: uncharacterized protein LOC105643225 [Jatropha curcas] |