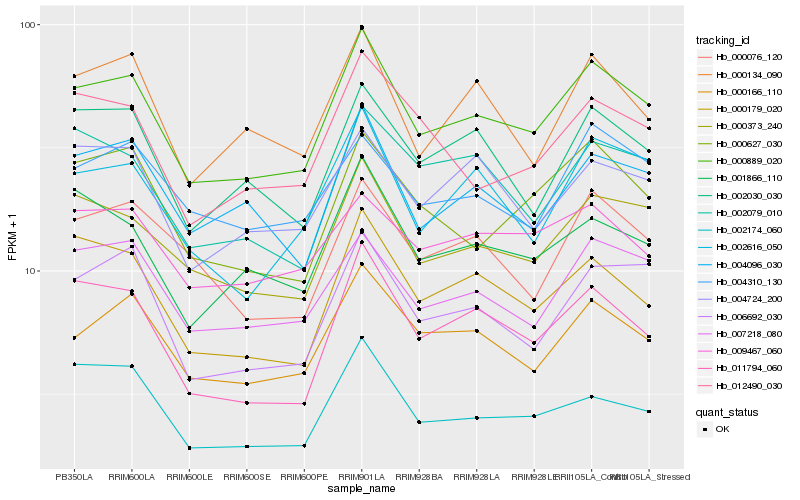
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000889_020 |
0.0 |
- |
- |
ribosome biogenesis protein brix, putative [Ricinus communis] |
| 2 |
Hb_000373_240 |
0.0597016304 |
- |
- |
PREDICTED: uncharacterized protein LOC105640940 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000166_110 |
0.0615505849 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 25 [Jatropha curcas] |
| 4 |
Hb_007218_080 |
0.0692018677 |
- |
- |
PREDICTED: uncharacterized protein LOC105631523 [Jatropha curcas] |
| 5 |
Hb_011794_060 |
0.0715526796 |
- |
- |
PREDICTED: beta-amylase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000179_020 |
0.0722703866 |
- |
- |
amidase, putative [Ricinus communis] |
| 7 |
Hb_006692_030 |
0.0747333621 |
- |
- |
PREDICTED: uncharacterized protein LOC105632913 [Jatropha curcas] |
| 8 |
Hb_004096_030 |
0.0748615758 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002616_050 |
0.075004452 |
- |
- |
PREDICTED: traB domain-containing protein [Jatropha curcas] |
| 10 |
Hb_001866_110 |
0.0762794476 |
- |
- |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 11 |
Hb_002030_030 |
0.0782266316 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 12 |
Hb_000627_030 |
0.0789591779 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 13 |
Hb_000134_090 |
0.0805080437 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002174_060 |
0.0807004836 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 15 |
Hb_004724_200 |
0.0814298917 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 16 |
Hb_000076_120 |
0.0816935009 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 17 |
Hb_012490_030 |
0.0828032589 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004310_130 |
0.0833841708 |
- |
- |
PREDICTED: uncharacterized protein LOC105643225 [Jatropha curcas] |
| 19 |
Hb_002079_010 |
0.0834975142 |
- |
- |
hypothetical protein POPTR_0001s01970g [Populus trichocarpa] |
| 20 |
Hb_009467_060 |
0.0845694243 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |