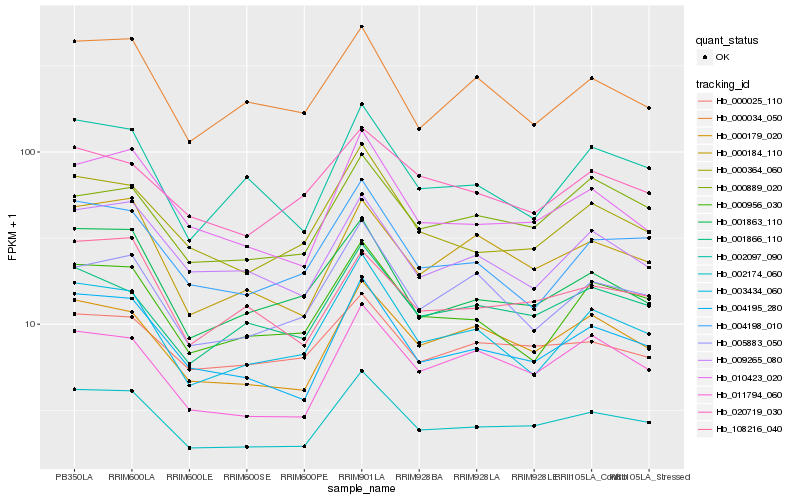
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002174_060 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 2 |
Hb_004195_280 |
0.0504477477 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 3 |
Hb_000179_020 |
0.0628839407 |
- |
- |
amidase, putative [Ricinus communis] |
| 4 |
Hb_000364_060 |
0.0680468578 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 5 |
Hb_010423_020 |
0.0685733526 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 6 |
Hb_001866_110 |
0.0738312478 |
- |
- |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 7 |
Hb_000184_110 |
0.076176046 |
- |
- |
hypothetical protein F383_18769 [Gossypium arboreum] |
| 8 |
Hb_011794_060 |
0.076810457 |
- |
- |
PREDICTED: beta-amylase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000025_110 |
0.0771864375 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 10 |
Hb_003434_060 |
0.0772909988 |
- |
- |
PREDICTED: membrane-bound transcription factor site-2 protease homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_001863_110 |
0.0789241861 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009265_080 |
0.0791761778 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_000956_030 |
0.0800637183 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 14 |
Hb_000889_020 |
0.0807004836 |
- |
- |
ribosome biogenesis protein brix, putative [Ricinus communis] |
| 15 |
Hb_004198_010 |
0.0821244257 |
- |
- |
Eukaryotic translation initiation factor 2 subunit beta [Theobroma cacao] |
| 16 |
Hb_108216_040 |
0.0846262341 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 17 |
Hb_005883_050 |
0.0849294206 |
- |
- |
PREDICTED: transportin-3 [Jatropha curcas] |
| 18 |
Hb_002097_090 |
0.0872094535 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 19 |
Hb_020719_030 |
0.0876059898 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 20 |
Hb_000034_050 |
0.0878660433 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |