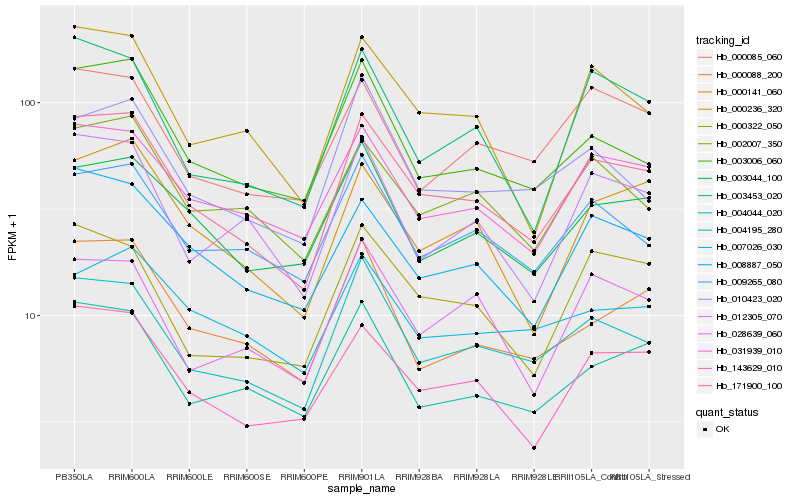
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171900_100 |
0.0 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0270496 [Jatropha curcas] |
| 2 |
Hb_004195_280 |
0.0678510834 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 3 |
Hb_143629_010 |
0.06918758 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 4 |
Hb_031939_010 |
0.0752820192 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_008887_050 |
0.0763135706 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 6 |
Hb_000236_320 |
0.0820068466 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 7 |
Hb_002007_350 |
0.0823339899 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 8 |
Hb_003006_060 |
0.0828434177 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 9 |
Hb_000141_060 |
0.0829945701 |
- |
- |
PREDICTED: uncharacterized protein LOC105632248 [Jatropha curcas] |
| 10 |
Hb_009265_080 |
0.0844897537 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 11 |
Hb_000322_050 |
0.0849070097 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_003044_100 |
0.0859545499 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 13 |
Hb_003453_020 |
0.0870771666 |
- |
- |
PREDICTED: histidine biosynthesis bifunctional protein hisIE, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004044_020 |
0.0887173107 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000088_200 |
0.0916270078 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_012305_070 |
0.0917095113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000085_060 |
0.0918771443 |
- |
- |
PREDICTED: uncharacterized protein LOC105633020 [Jatropha curcas] |
| 18 |
Hb_028639_060 |
0.0944779318 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_010423_020 |
0.0953189324 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 20 |
Hb_007026_030 |
0.095468596 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |