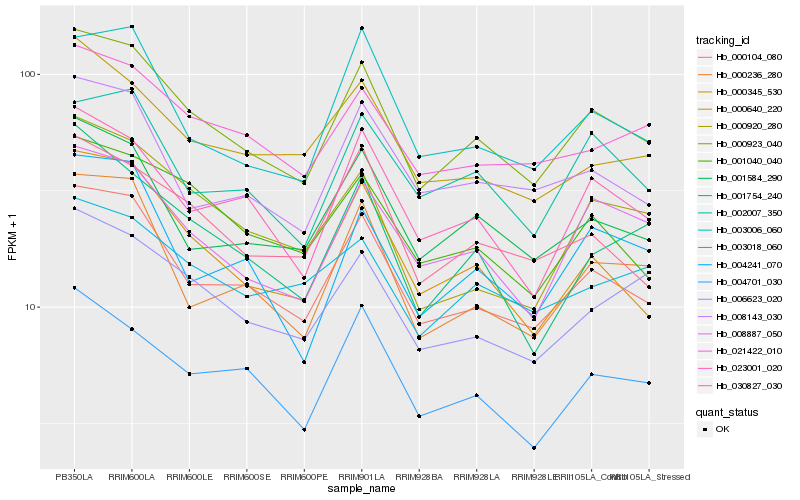
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000923_040 |
0.0 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 2 |
Hb_000104_080 |
0.0458646187 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 3 |
Hb_008887_050 |
0.0676482133 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 4 |
Hb_000345_530 |
0.0702726498 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001584_290 |
0.0708485593 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_023001_020 |
0.0709424928 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 7 |
Hb_001040_040 |
0.073825853 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 8 |
Hb_030827_030 |
0.0780435856 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 9 |
Hb_021422_010 |
0.0788128777 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004701_030 |
0.0790505952 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 11 |
Hb_006623_020 |
0.0796266114 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003006_060 |
0.080679311 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 13 |
Hb_000236_280 |
0.0808926016 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_002007_350 |
0.0825381547 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 15 |
Hb_000640_220 |
0.0842819545 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 16 |
Hb_000920_280 |
0.0855168202 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 17 |
Hb_008143_030 |
0.0855946191 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004241_070 |
0.0863034676 |
- |
- |
PREDICTED: WD repeat-containing protein 25 [Jatropha curcas] |
| 19 |
Hb_003018_060 |
0.0880206618 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 20 |
Hb_001754_240 |
0.0880910171 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |