| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
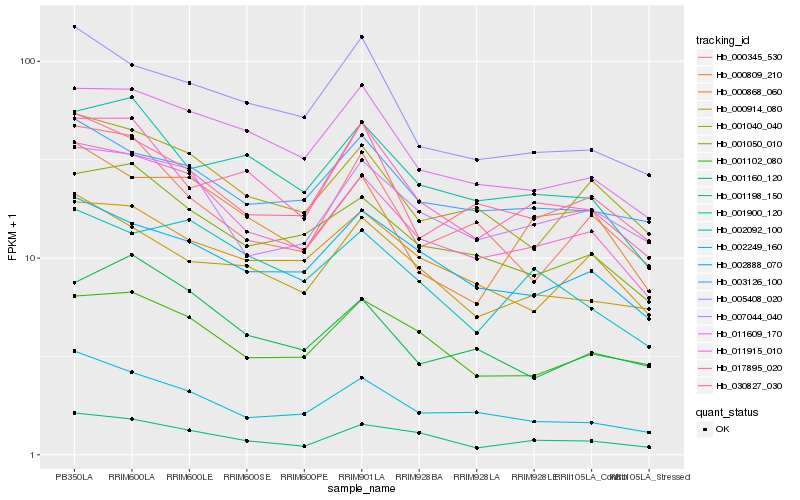
Hb_011915_010 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 2 |
Hb_001040_040 |
0.072354767 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 3 |
Hb_001050_010 |
0.0747024395 |
- |
- |
Advillin [Gossypium arboreum] |
| 4 |
Hb_002249_160 |
0.0753038109 |
- |
- |
- |
| 5 |
Hb_011609_170 |
0.0819737236 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 6 |
Hb_007044_040 |
0.0845464718 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 7 |
Hb_001198_150 |
0.0891755095 |
- |
- |
hypothetical protein POPTR_0003s01280g [Populus trichocarpa] |
| 8 |
Hb_002888_070 |
0.0907713009 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 9 |
Hb_030827_030 |
0.0928001 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 10 |
Hb_001102_080 |
0.0946105211 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 11 |
Hb_000868_060 |
0.0954537921 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_000809_210 |
0.0957800412 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 13 |
Hb_001160_120 |
0.0964614065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003126_100 |
0.0969889806 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 15 |
Hb_001900_120 |
0.0986890101 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 16 |
Hb_005408_020 |
0.0988951549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000914_080 |
0.0995362742 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_002092_100 |
0.0998996933 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 19 |
Hb_000345_530 |
0.1004125788 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 20 |
Hb_017895_020 |
0.1021487837 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |