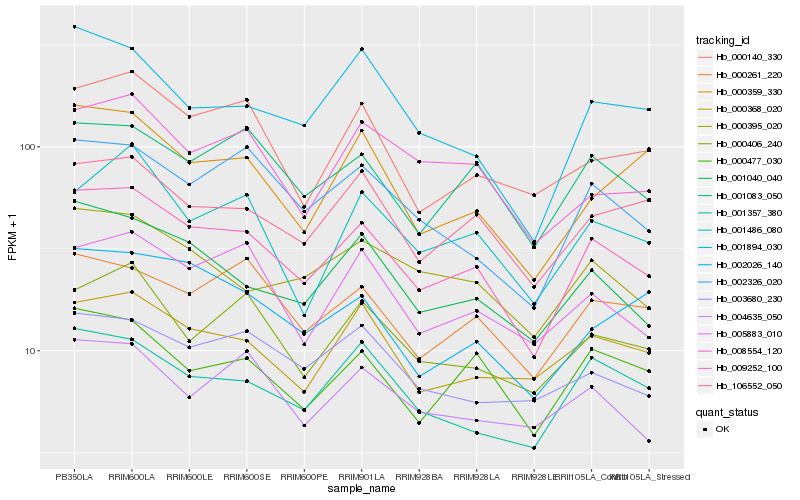
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009252_100 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 2 |
Hb_001040_040 |
0.0768394409 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 3 |
Hb_001083_050 |
0.0831051242 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 4 |
Hb_106552_050 |
0.0842364498 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 5 |
Hb_000477_030 |
0.0843587904 |
- |
- |
PREDICTED: boron transporter 4-like [Jatropha curcas] |
| 6 |
Hb_002326_020 |
0.0848677814 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 7 |
Hb_001357_380 |
0.0858682017 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_000261_220 |
0.0860359777 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 9 |
Hb_000406_240 |
0.0899095016 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_005883_010 |
0.0902910692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000395_020 |
0.0907777003 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 12 |
Hb_000368_020 |
0.0920028061 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000140_330 |
0.0958025424 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 14 |
Hb_008554_120 |
0.0958493553 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 15 |
Hb_001894_030 |
0.0963138661 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 16 |
Hb_002026_140 |
0.0981179872 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 17 |
Hb_001486_080 |
0.0985258023 |
- |
- |
PREDICTED: eukaryotic translation initiation factor NCBP [Jatropha curcas] |
| 18 |
Hb_003680_230 |
0.0986565704 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 19 |
Hb_000359_330 |
0.102075818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004635_050 |
0.1023753532 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |