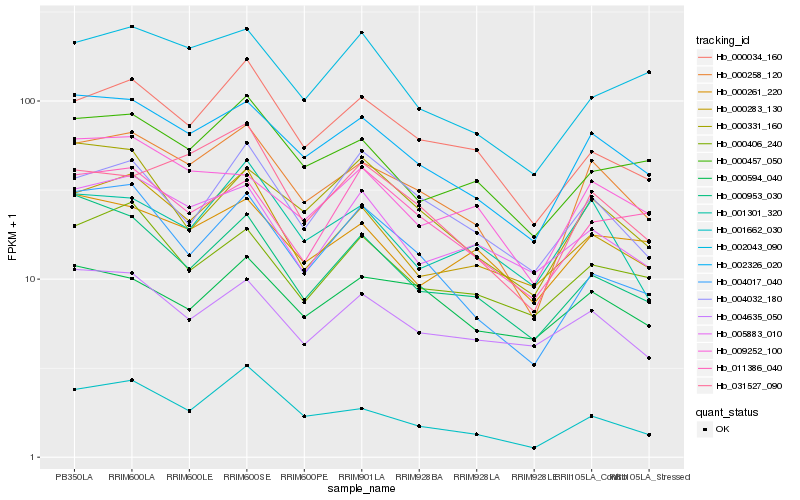
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 2 |
Hb_002326_020 |
0.0627636473 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 3 |
Hb_000034_160 |
0.0975770773 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 4 |
Hb_001662_030 |
0.1008066969 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 5 |
Hb_031527_090 |
0.1018091396 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000283_130 |
0.1041465409 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 7 |
Hb_000457_050 |
0.108688286 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 8 |
Hb_009252_100 |
0.1135880596 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 9 |
Hb_001301_320 |
0.1148656761 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 10 |
Hb_002043_090 |
0.1168443776 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 11 |
Hb_000953_030 |
0.118160881 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000406_240 |
0.1185524622 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 13 |
Hb_004017_040 |
0.1191641726 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 14 |
Hb_004635_050 |
0.1198832028 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_005883_010 |
0.1200645858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004032_180 |
0.1200744184 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 17 |
Hb_000261_220 |
0.120275695 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 18 |
Hb_000331_160 |
0.1212354249 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 19 |
Hb_000594_040 |
0.1214045449 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 20 |
Hb_011386_040 |
0.1215586626 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |