| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
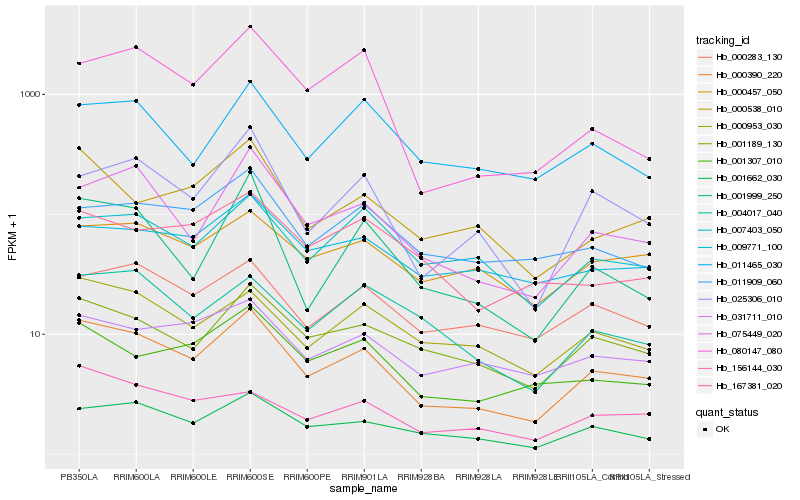
Hb_000390_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 2 |
Hb_001662_030 |
0.1039111067 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 3 |
Hb_075449_020 |
0.1159442878 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 4 |
Hb_001189_130 |
0.1170826511 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 5 |
Hb_025306_010 |
0.1226246664 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |
| 6 |
Hb_001307_010 |
0.1308507336 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 7 |
Hb_000953_030 |
0.131111141 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000538_010 |
0.1336362977 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000283_130 |
0.1352347932 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 10 |
Hb_156144_030 |
0.1354908831 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 11 |
Hb_031711_010 |
0.1386866103 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001999_250 |
0.1404788302 |
- |
- |
PREDICTED: uncharacterized protein LOC105133073 isoform X1 [Populus euphratica] |
| 13 |
Hb_004017_040 |
0.1410421703 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 14 |
Hb_000457_050 |
0.1414102054 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 15 |
Hb_011465_030 |
0.143438736 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 16 |
Hb_007403_050 |
0.1436053901 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 17 |
Hb_009771_100 |
0.1453229368 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 18 |
Hb_011909_060 |
0.1459777821 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 19 |
Hb_167381_020 |
0.1492884064 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 20 |
Hb_080147_080 |
0.1503564744 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |