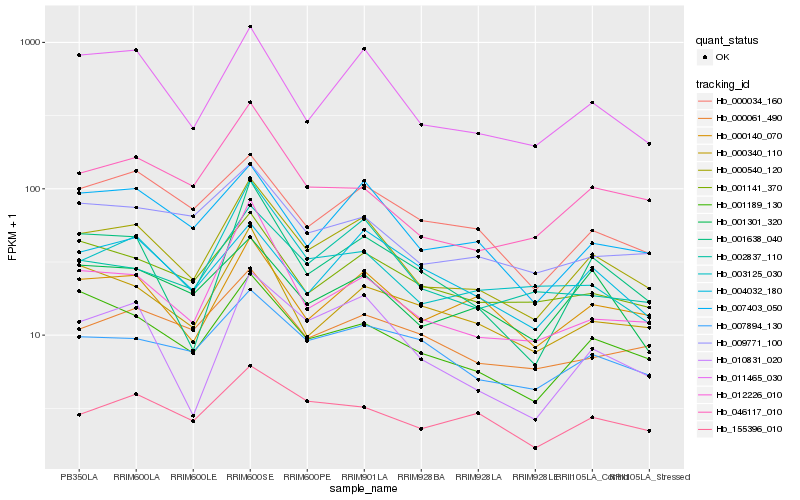
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_120 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 2 |
Hb_011465_030 |
0.090893144 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_007403_050 |
0.0979923442 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 4 |
Hb_001189_130 |
0.10104034 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 5 |
Hb_000140_070 |
0.1028594124 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010831_020 |
0.1086590131 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_003125_030 |
0.1090957971 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 8 |
Hb_001638_040 |
0.1095001676 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 9 |
Hb_012226_010 |
0.1098187254 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 10 |
Hb_007894_130 |
0.1104979283 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 11 |
Hb_000034_160 |
0.1115886107 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 12 |
Hb_001141_370 |
0.1127917049 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 13 |
Hb_002837_110 |
0.1142109804 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_009771_100 |
0.115687142 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 15 |
Hb_155396_010 |
0.1158859318 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 16 |
Hb_000061_490 |
0.1172716781 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 17 |
Hb_046117_010 |
0.1175732999 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 18 |
Hb_001301_320 |
0.1182649266 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 19 |
Hb_000340_110 |
0.1183969761 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 20 |
Hb_004032_180 |
0.118957066 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |