| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
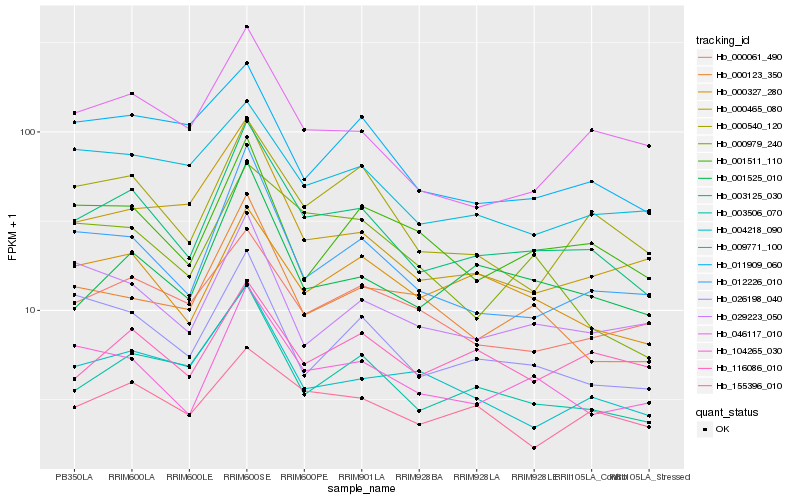
Hb_003125_030 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 2 |
Hb_104265_030 |
0.0984141964 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 3 |
Hb_012226_010 |
0.1006008977 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 4 |
Hb_003506_070 |
0.1015966714 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000540_120 |
0.1090957971 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 6 |
Hb_001525_010 |
0.1141069712 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 7 |
Hb_000327_280 |
0.1185462818 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 8 |
Hb_046117_010 |
0.1192578981 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 9 |
Hb_001511_110 |
0.1229727671 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_026198_040 |
0.1235304822 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 11 |
Hb_000123_350 |
0.1257480659 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_004218_090 |
0.1269883977 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_155396_010 |
0.1280155343 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 14 |
Hb_000061_490 |
0.1305286086 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 15 |
Hb_009771_100 |
0.1316431155 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 16 |
Hb_000465_080 |
0.1318349999 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 17 |
Hb_029223_050 |
0.1338152999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 18 |
Hb_116086_010 |
0.1345635265 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 19 |
Hb_011909_060 |
0.1347015276 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 20 |
Hb_000979_240 |
0.1365711783 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |