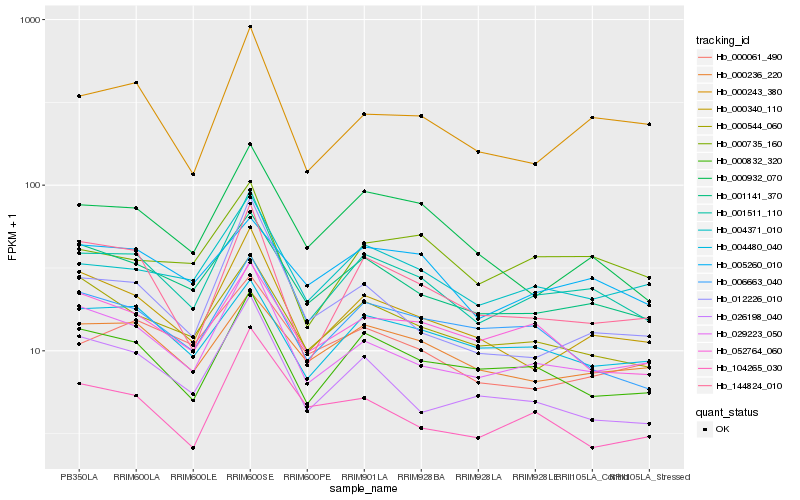
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_110 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_029223_050 |
0.0777766166 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000340_110 |
0.0799898201 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 4 |
Hb_001141_370 |
0.0860890202 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 5 |
Hb_000832_320 |
0.0873920124 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_000243_380 |
0.0894102017 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 7 |
Hb_004371_010 |
0.0901860961 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_144824_010 |
0.0959728031 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 9 |
Hb_000544_060 |
0.0977588003 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 10 |
Hb_026198_040 |
0.1047434317 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 11 |
Hb_000932_070 |
0.1051926125 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 12 |
Hb_006663_040 |
0.1060919434 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_012226_010 |
0.1064153659 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 14 |
Hb_004480_040 |
0.107046149 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000236_220 |
0.109355497 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 16 |
Hb_052764_060 |
0.109984849 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000735_160 |
0.1110519506 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 18 |
Hb_104265_030 |
0.1116036702 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 19 |
Hb_000061_490 |
0.1143870881 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 20 |
Hb_005260_010 |
0.1144764565 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |