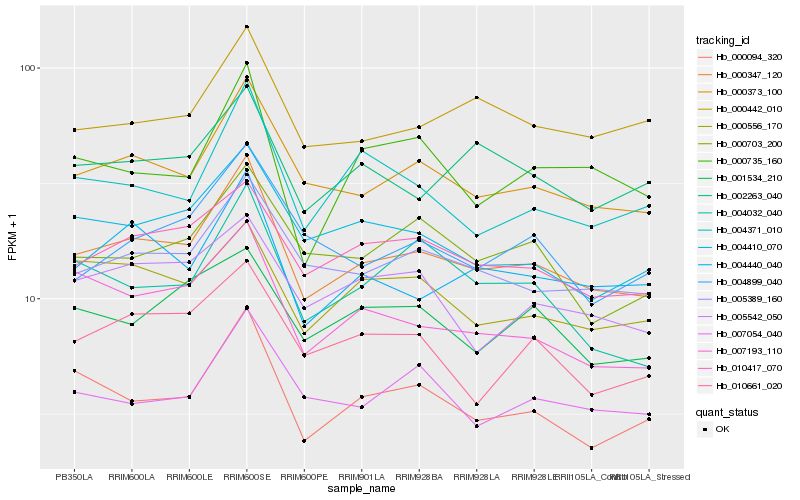
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 2 |
Hb_000373_100 |
0.0528633081 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 3 |
Hb_005389_160 |
0.0677485342 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 4 |
Hb_010417_070 |
0.0739359848 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004440_040 |
0.0762182737 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000094_320 |
0.0780734968 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 7 |
Hb_004410_070 |
0.0802328762 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 8 |
Hb_007193_110 |
0.0841009195 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 9 |
Hb_000442_010 |
0.0860353419 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 10 |
Hb_000556_170 |
0.0868792055 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004371_010 |
0.0873336415 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_007054_040 |
0.0886066809 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 13 |
Hb_002263_040 |
0.0886949461 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 14 |
Hb_000703_200 |
0.0910650367 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_005542_050 |
0.0921405846 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 16 |
Hb_010661_020 |
0.0924567619 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000735_160 |
0.0943835244 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 18 |
Hb_004899_040 |
0.0950683756 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 19 |
Hb_001534_210 |
0.0959931222 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004032_040 |
0.0968789655 |
- |
- |
unknown [Medicago truncatula] |