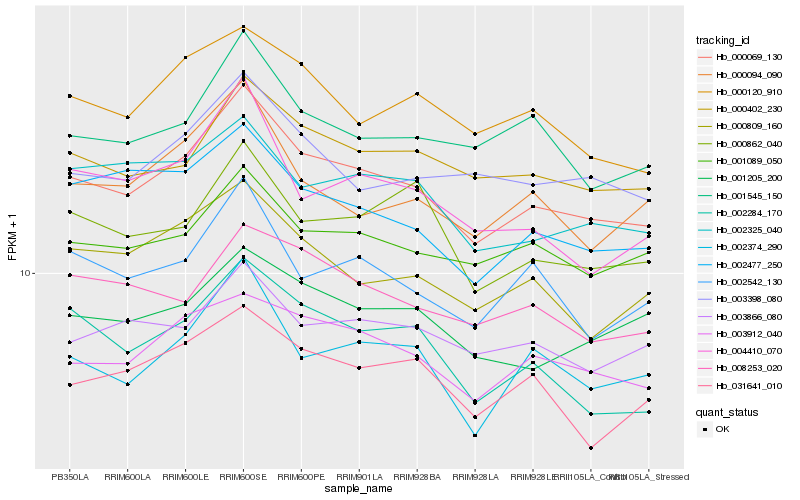
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003912_040 |
0.0589202218 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 3 |
Hb_000402_230 |
0.0602169074 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000809_160 |
0.0656916473 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 5 |
Hb_001089_050 |
0.065831039 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_002374_290 |
0.0674670049 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002542_130 |
0.0678738779 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 8 |
Hb_000120_910 |
0.0681134594 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 9 |
Hb_002477_250 |
0.0682293389 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 10 |
Hb_001545_150 |
0.0682302394 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 11 |
Hb_003398_080 |
0.0687328614 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_002325_040 |
0.06982963 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008253_020 |
0.0702107613 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 14 |
Hb_001205_200 |
0.0702401942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002284_170 |
0.071170164 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 16 |
Hb_003866_080 |
0.0711778987 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 17 |
Hb_000094_090 |
0.0719329311 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 18 |
Hb_031641_010 |
0.072084418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004410_070 |
0.0722230982 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 20 |
Hb_000862_040 |
0.0728507166 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |