| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
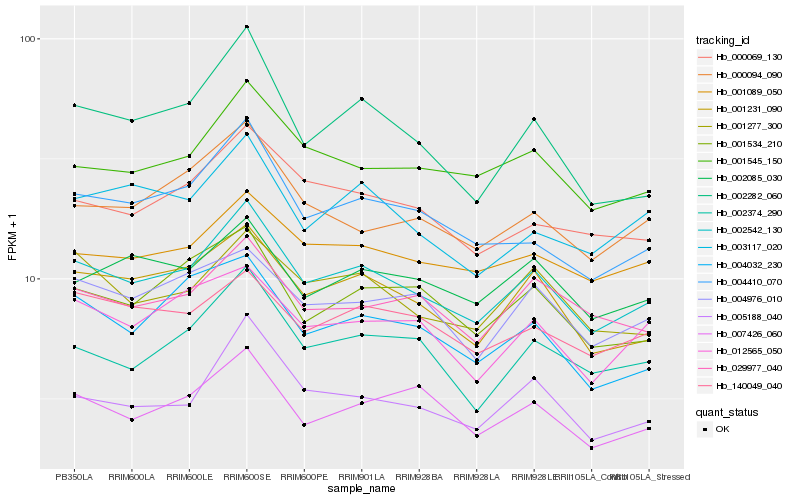
Hb_002542_130 |
0.0 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 2 |
Hb_001231_090 |
0.0532512471 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001545_150 |
0.0638628565 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 4 |
Hb_001277_300 |
0.0651957637 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 5 |
Hb_005188_040 |
0.0658895322 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 6 |
Hb_004410_070 |
0.0665164954 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 7 |
Hb_002282_060 |
0.067638109 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_000069_130 |
0.0678738779 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001089_050 |
0.0681721795 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_002374_290 |
0.0682050441 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001534_210 |
0.068614711 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004976_010 |
0.0689320466 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 13 |
Hb_140049_040 |
0.0701184286 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 14 |
Hb_007426_060 |
0.070569562 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_029977_040 |
0.0705856651 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 16 |
Hb_000094_090 |
0.0713473641 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012565_050 |
0.0713749142 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein 70 kDa [Jatropha curcas] |
| 18 |
Hb_002085_030 |
0.0718886999 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003117_020 |
0.0733219643 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 20 |
Hb_004032_230 |
0.0754242836 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |