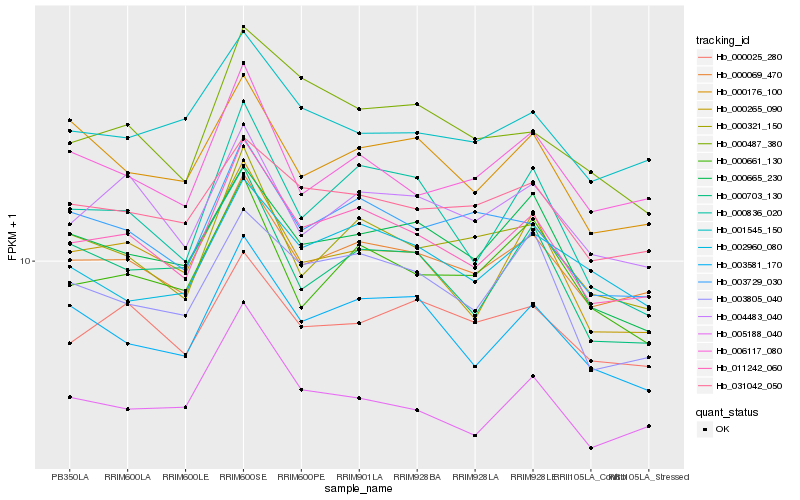
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011242_060 |
0.0 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 2 |
Hb_000069_470 |
0.0458698467 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003581_170 |
0.0600412252 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000265_090 |
0.064080928 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 5 |
Hb_003729_030 |
0.0651578704 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 6 |
Hb_000836_020 |
0.0674429816 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 7 |
Hb_000321_150 |
0.0692893227 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004483_040 |
0.0693981298 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 9 |
Hb_005188_040 |
0.0695969936 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 10 |
Hb_003805_040 |
0.0701061234 |
- |
- |
Sacsin [Glycine soja] |
| 11 |
Hb_031042_050 |
0.0718280539 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_000665_230 |
0.0727072484 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_000487_380 |
0.0731589 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006117_080 |
0.07530144 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000025_280 |
0.0772155901 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 16 |
Hb_000176_100 |
0.0772395041 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 17 |
Hb_000703_130 |
0.0774379587 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 18 |
Hb_001545_150 |
0.0795372614 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 19 |
Hb_002960_080 |
0.0796354288 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 20 |
Hb_000661_130 |
0.0797589952 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |