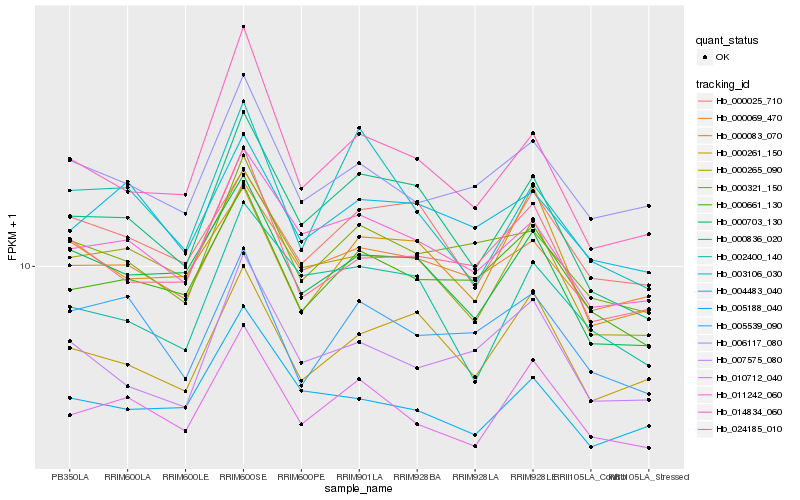
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010712_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290 [Jatropha curcas] |
| 2 |
Hb_000661_130 |
0.0715540174 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 3 |
Hb_000265_090 |
0.0722669536 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 4 |
Hb_011242_060 |
0.0816045117 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 5 |
Hb_006117_080 |
0.085608094 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000703_130 |
0.0876098362 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 7 |
Hb_000836_020 |
0.0885031149 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 8 |
Hb_003106_030 |
0.0924128311 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 9 |
Hb_000069_470 |
0.0932901795 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005188_040 |
0.0950989991 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 11 |
Hb_004483_040 |
0.0955523774 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 12 |
Hb_005539_090 |
0.0971079698 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 13 |
Hb_024185_010 |
0.0986198023 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_014834_060 |
0.1001699089 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000083_070 |
0.1004789904 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002400_140 |
0.1017696779 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 17 |
Hb_000321_150 |
0.102059166 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000261_150 |
0.1024262561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007575_080 |
0.1033107023 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 20 |
Hb_000025_710 |
0.1036739327 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |