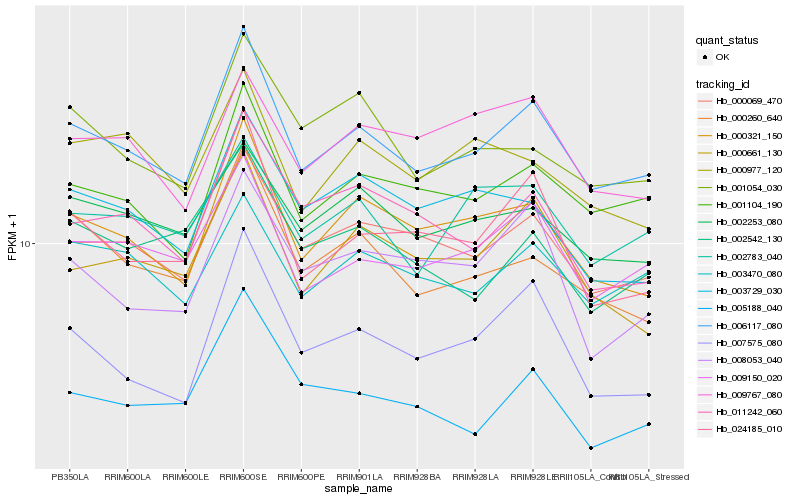
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006117_080 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001104_190 |
0.0546116039 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 3 |
Hb_000069_470 |
0.0548372589 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009150_020 |
0.0554653313 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 5 |
Hb_000321_150 |
0.0561396629 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 6 |
Hb_024185_010 |
0.0646951367 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000260_640 |
0.0683860353 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 8 |
Hb_003470_080 |
0.0705317231 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000661_130 |
0.0712323471 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 10 |
Hb_002253_080 |
0.0715049137 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 11 |
Hb_007575_080 |
0.0719360091 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 12 |
Hb_011242_060 |
0.07530144 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 13 |
Hb_002783_040 |
0.0756878653 |
transcription factor |
TF Family: IWS1 |
- |
| 14 |
Hb_001054_030 |
0.0777637765 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 15 |
Hb_005188_040 |
0.0788163326 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 16 |
Hb_003729_030 |
0.079845544 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 17 |
Hb_008053_040 |
0.0824112335 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 18 |
Hb_002542_130 |
0.082634012 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 19 |
Hb_000977_120 |
0.0826781127 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_009767_080 |
0.0840731624 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |