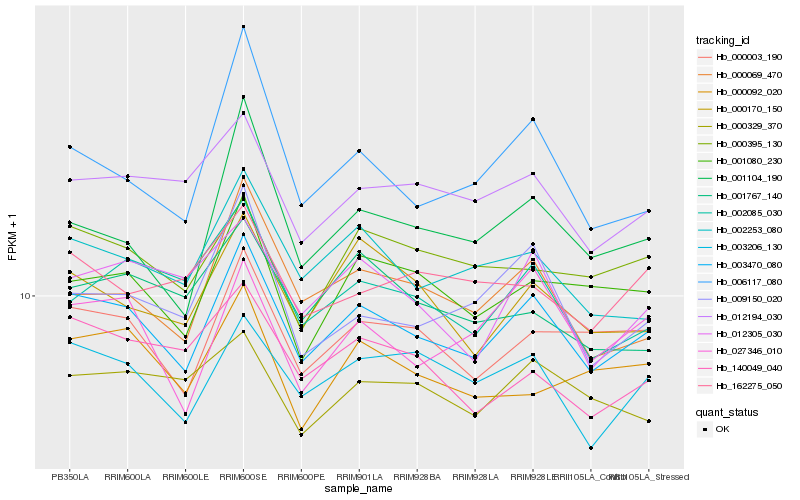
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003470_080 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000003_190 |
0.0535135706 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 3 |
Hb_003206_130 |
0.0557626131 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 4 |
Hb_002253_080 |
0.0603857923 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 5 |
Hb_001080_230 |
0.0619894917 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 6 |
Hb_000069_470 |
0.0628246483 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000170_150 |
0.0632784213 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 8 |
Hb_012194_030 |
0.0638200517 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001104_190 |
0.0655160774 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 10 |
Hb_027346_010 |
0.0668698363 |
- |
- |
- |
| 11 |
Hb_000395_130 |
0.0676128693 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 12 |
Hb_009150_020 |
0.0685819143 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 13 |
Hb_002085_030 |
0.0687871915 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_140049_040 |
0.069426551 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 15 |
Hb_012305_030 |
0.0697969772 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 16 |
Hb_006117_080 |
0.0705317231 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000092_020 |
0.0715114131 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000329_370 |
0.073067577 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 19 |
Hb_001767_140 |
0.0757394362 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 20 |
Hb_162275_050 |
0.0759896464 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |