| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
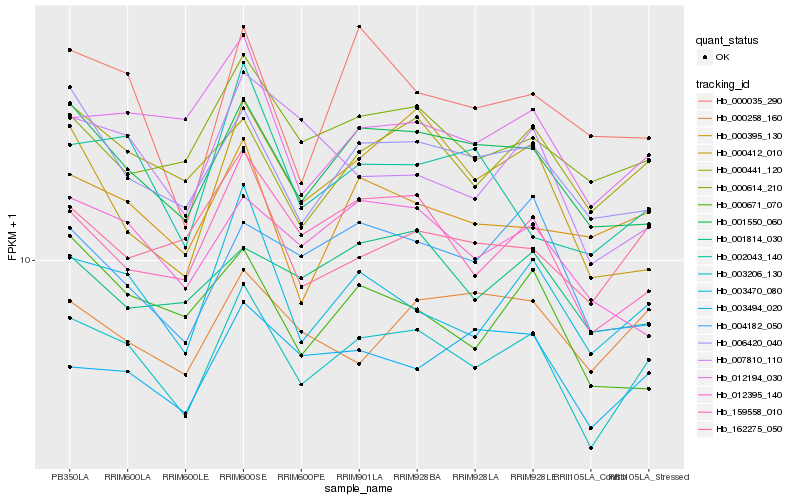
Hb_003206_130 |
0.0 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 2 |
Hb_001550_060 |
0.0541756688 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 3 |
Hb_003470_080 |
0.0557626131 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000441_120 |
0.0582031919 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_159558_010 |
0.0584472644 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 6 |
Hb_006420_040 |
0.0599292286 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000614_210 |
0.0632157443 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003494_020 |
0.0653238241 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 9 |
Hb_012194_030 |
0.0654254912 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_162275_050 |
0.0685357253 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 11 |
Hb_000395_130 |
0.0689660794 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 12 |
Hb_000258_160 |
0.070695225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012395_140 |
0.0718322055 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 14 |
Hb_004182_050 |
0.0726332118 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001814_030 |
0.0727065023 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 16 |
Hb_007810_110 |
0.0761203358 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 17 |
Hb_000035_290 |
0.0765372035 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 18 |
Hb_002043_140 |
0.0768159451 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 19 |
Hb_000671_070 |
0.0768928013 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 20 |
Hb_000412_010 |
0.0770607039 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |