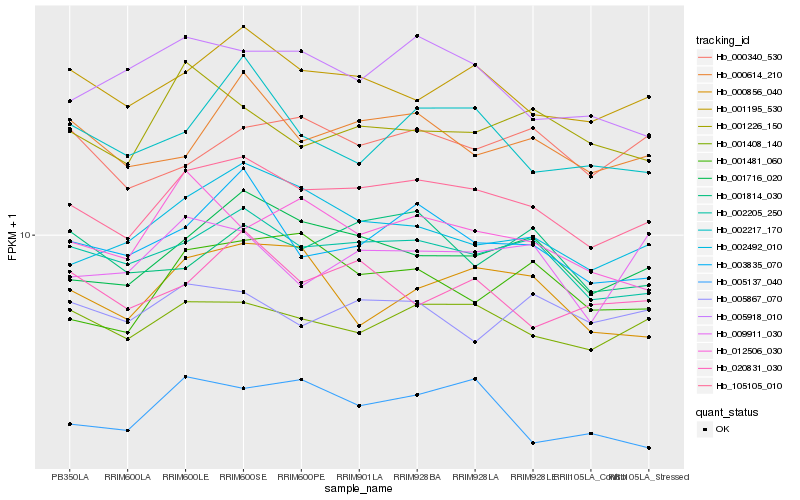
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105105_010 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 2 |
Hb_002205_250 |
0.0404822665 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001408_140 |
0.0462292205 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 4 |
Hb_001195_530 |
0.0487631476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001716_020 |
0.0492518249 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 6 |
Hb_003835_070 |
0.0533957149 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002492_010 |
0.0575960974 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_000614_210 |
0.059151558 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005137_040 |
0.060437439 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 10 |
Hb_001481_060 |
0.0641750797 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001226_150 |
0.0645204186 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_012506_030 |
0.0647293972 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 13 |
Hb_000856_040 |
0.0649727169 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 14 |
Hb_020831_030 |
0.0649944031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005918_010 |
0.0652803021 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 16 |
Hb_009911_030 |
0.0663068638 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 17 |
Hb_002217_170 |
0.0672264824 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 18 |
Hb_005867_070 |
0.0682703693 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000340_530 |
0.0684028221 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 20 |
Hb_001814_030 |
0.0684162894 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |