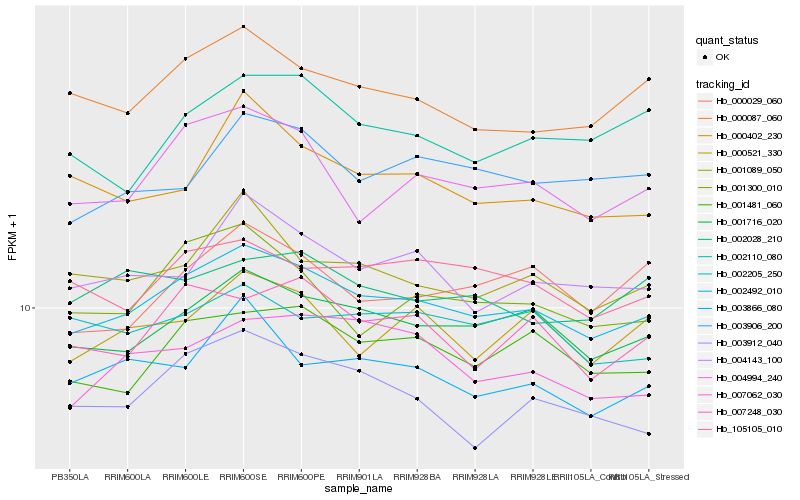
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002492_010 |
0.0 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 2 |
Hb_001716_020 |
0.0364425602 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 3 |
Hb_001481_060 |
0.0513735423 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000087_060 |
0.05484306 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 5 |
Hb_004994_240 |
0.0557864243 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 6 |
Hb_001089_050 |
0.0564011205 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_004143_100 |
0.057011576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_105105_010 |
0.0575960974 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 9 |
Hb_007248_030 |
0.0596397142 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 10 |
Hb_003866_080 |
0.0602811062 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 11 |
Hb_000521_330 |
0.060415723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001300_010 |
0.0606669361 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_003906_200 |
0.0610177708 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 14 |
Hb_003912_040 |
0.0625551137 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 15 |
Hb_007062_030 |
0.0635822955 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 16 |
Hb_000029_060 |
0.064717216 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 17 |
Hb_002028_210 |
0.0647187426 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 18 |
Hb_002205_250 |
0.0648816408 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002110_080 |
0.0649694384 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 20 |
Hb_000402_230 |
0.0656241699 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |