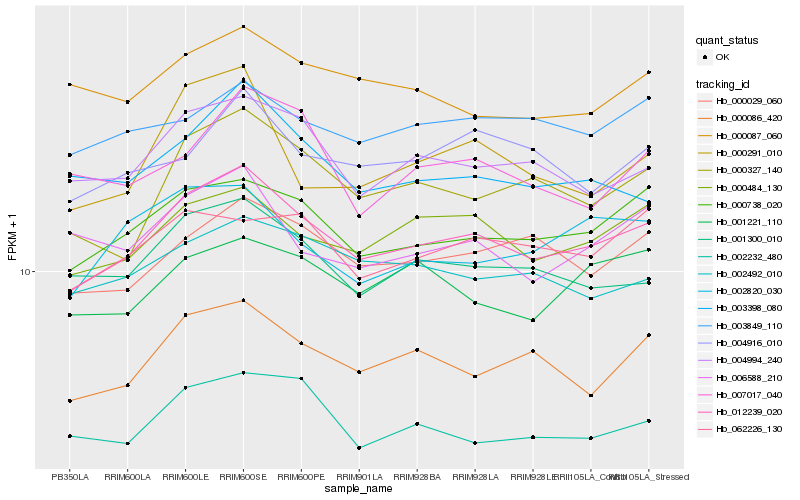
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012239_020 |
0.0 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 2 |
Hb_000738_020 |
0.0556124831 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000484_130 |
0.0623332385 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 4 |
Hb_000086_420 |
0.0636182839 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 5 |
Hb_004994_240 |
0.0642258151 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 6 |
Hb_001300_010 |
0.0648261538 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000327_140 |
0.0667836363 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000029_060 |
0.0673276925 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 9 |
Hb_002492_010 |
0.0734094948 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 10 |
Hb_002232_480 |
0.0748707482 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 11 |
Hb_003398_080 |
0.0754690096 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_062226_130 |
0.0765754352 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 13 |
Hb_004916_010 |
0.0781002656 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 14 |
Hb_006588_210 |
0.0786424153 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 15 |
Hb_001221_110 |
0.078920789 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 16 |
Hb_002820_030 |
0.0789326804 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 17 |
Hb_003849_110 |
0.0792754949 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 18 |
Hb_000087_060 |
0.0800267471 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_000291_010 |
0.080399356 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 20 |
Hb_007017_040 |
0.084558826 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |