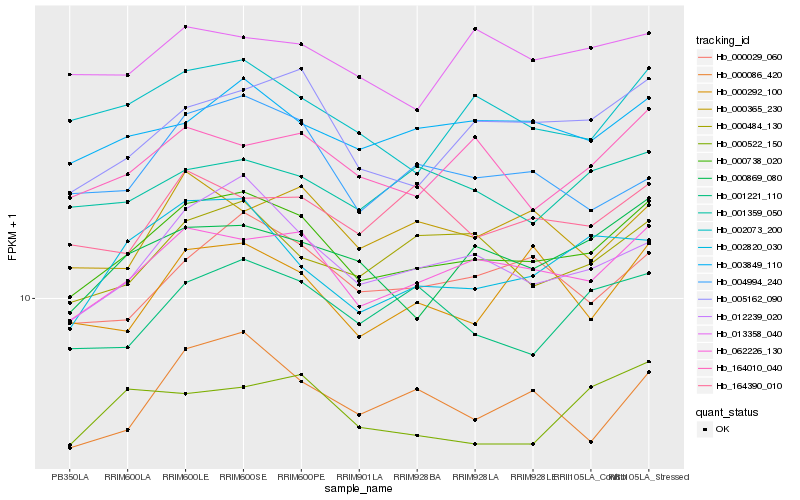
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000738_020 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_062226_130 |
0.0399179371 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 3 |
Hb_012239_020 |
0.0556124831 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 4 |
Hb_002073_200 |
0.0635943796 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 5 |
Hb_005162_090 |
0.0636561922 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 6 |
Hb_003849_110 |
0.0642415586 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 7 |
Hb_000869_080 |
0.0659912331 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000029_060 |
0.0669422015 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 9 |
Hb_164010_040 |
0.0673524849 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000484_130 |
0.0687879471 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 11 |
Hb_002820_030 |
0.0692309855 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 12 |
Hb_001221_110 |
0.0712290178 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 13 |
Hb_000086_420 |
0.0727473011 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 14 |
Hb_000522_150 |
0.0729427066 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 15 |
Hb_001359_050 |
0.0745017431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000365_230 |
0.0771759611 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 17 |
Hb_000292_100 |
0.0779007442 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 18 |
Hb_013358_040 |
0.078592199 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 19 |
Hb_164390_010 |
0.078680017 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 20 |
Hb_004994_240 |
0.078730156 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |