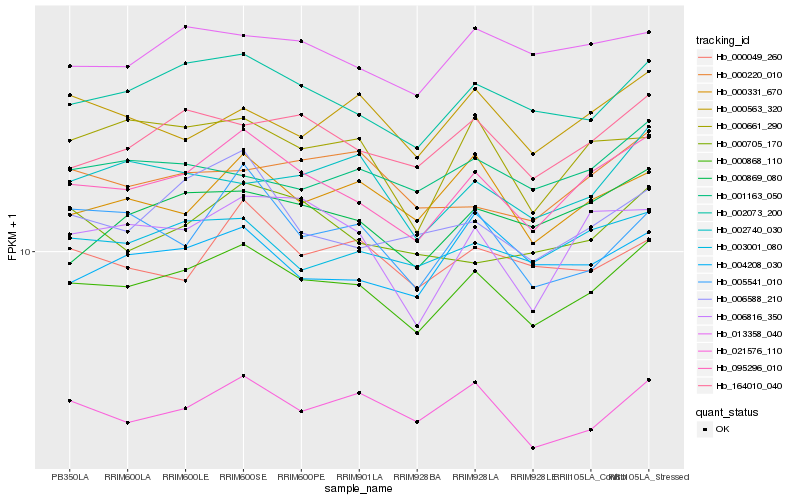
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000868_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 2 |
Hb_095296_010 |
0.0362758562 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 3 |
Hb_002073_200 |
0.051222627 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 4 |
Hb_164010_040 |
0.0657605065 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_003001_080 |
0.0697585473 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 6 |
Hb_021576_110 |
0.0698529266 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 7 |
Hb_000869_080 |
0.0737370895 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000331_670 |
0.0739435603 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 9 |
Hb_002740_030 |
0.074980207 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000661_290 |
0.0758166609 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_013358_040 |
0.0778996537 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 12 |
Hb_006588_210 |
0.0795104755 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 13 |
Hb_000049_260 |
0.0807046078 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_006816_350 |
0.080948521 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 15 |
Hb_001163_050 |
0.0809748975 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 16 |
Hb_000563_320 |
0.0810946899 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 17 |
Hb_000220_010 |
0.0813847931 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 18 |
Hb_000705_170 |
0.0814610503 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 19 |
Hb_005541_010 |
0.0817421408 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 20 |
Hb_004208_030 |
0.0829470411 |
transcription factor |
TF Family: MYB-related |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |