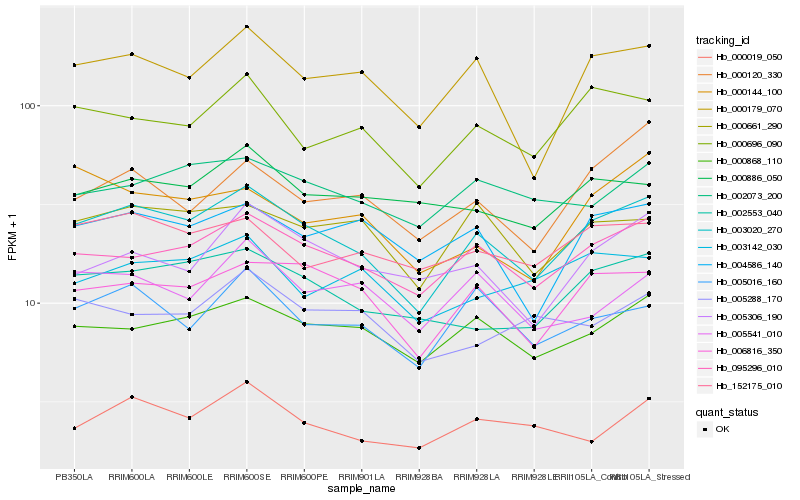
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_270 |
0.0 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 2 |
Hb_095296_010 |
0.0717845615 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 3 |
Hb_006816_350 |
0.0763590348 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |
| 4 |
Hb_000179_070 |
0.0818715882 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 5 |
Hb_000868_110 |
0.0854862232 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 6 |
Hb_002553_040 |
0.0862186676 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 7 |
Hb_000696_090 |
0.0879581242 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 8 |
Hb_000661_290 |
0.0916263297 |
- |
- |
PREDICTED: NAD(H) kinase 1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_005016_160 |
0.0925083635 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 10 |
Hb_004586_140 |
0.099075494 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 11 |
Hb_152175_010 |
0.0996266418 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 12 |
Hb_002073_200 |
0.1017387321 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 13 |
Hb_000019_050 |
0.1021133357 |
- |
- |
PREDICTED: protein NETWORKED 3A-like [Jatropha curcas] |
| 14 |
Hb_005306_190 |
0.1026121009 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 15 |
Hb_005541_010 |
0.1046175198 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 16 |
Hb_000120_330 |
0.1066800785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003142_030 |
0.1075639079 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000886_050 |
0.1080986787 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 19 |
Hb_005288_170 |
0.1115167345 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 20 |
Hb_000144_100 |
0.1126993036 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |