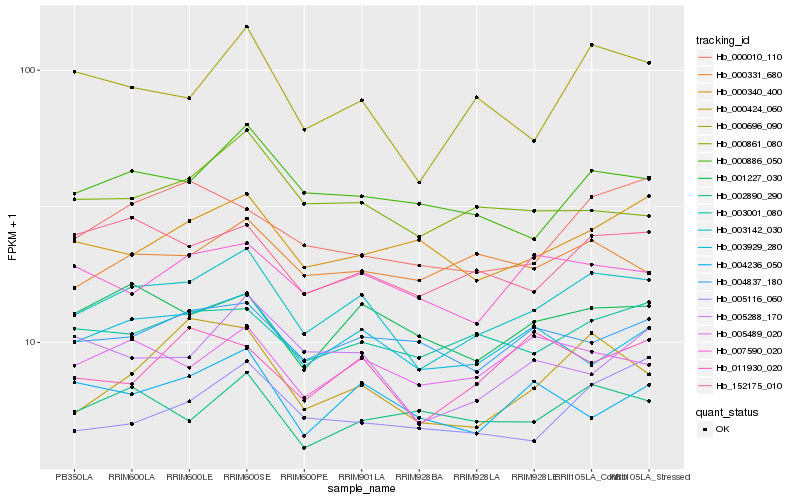
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003142_030 |
0.0 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000696_090 |
0.0772795513 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 3 |
Hb_152175_010 |
0.0787285689 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 4 |
Hb_003001_080 |
0.0790768274 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 5 |
Hb_000424_060 |
0.0793334031 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 6 |
Hb_001227_030 |
0.0803387341 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003929_280 |
0.0804258196 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 8 |
Hb_000886_050 |
0.0810834534 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 9 |
Hb_005489_020 |
0.0830102986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004837_180 |
0.0860334206 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000331_680 |
0.0865061913 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 12 |
Hb_007590_020 |
0.0865132805 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 13 |
Hb_000861_080 |
0.0881635204 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 14 |
Hb_002890_290 |
0.0885961265 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 15 |
Hb_005288_170 |
0.088665066 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 16 |
Hb_005116_060 |
0.0889890306 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000010_110 |
0.089554653 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004236_050 |
0.0903605214 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 19 |
Hb_000340_400 |
0.0907712029 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 20 |
Hb_011930_020 |
0.0913016395 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |