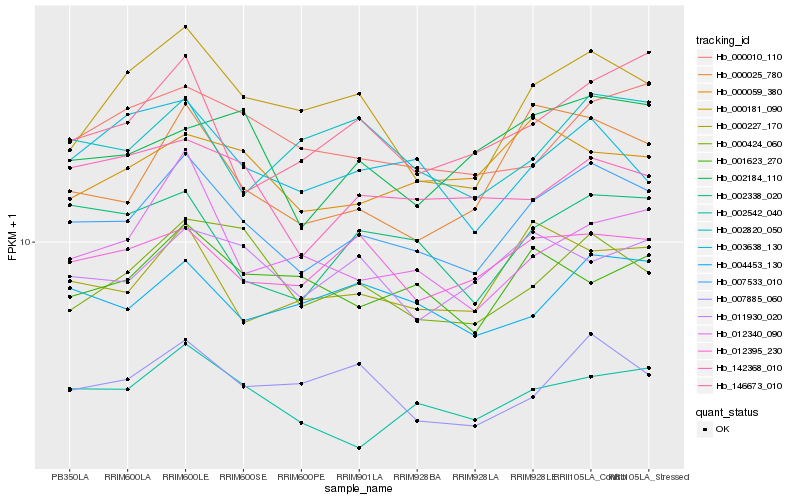
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007533_010 |
0.0 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 2 |
Hb_000025_780 |
0.0888573504 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 3 |
Hb_002338_020 |
0.0915786949 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 4 |
Hb_000227_170 |
0.0943326245 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_002542_040 |
0.0951125553 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 6 |
Hb_000181_090 |
0.0953377573 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 7 |
Hb_000424_060 |
0.0969010886 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 8 |
Hb_012395_230 |
0.0972426449 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 9 |
Hb_007885_060 |
0.0976841463 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 10 |
Hb_004453_130 |
0.0992637446 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |
| 11 |
Hb_000010_110 |
0.100286271 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_142368_010 |
0.1011808225 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_012340_090 |
0.102137498 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 14 |
Hb_003638_130 |
0.103511466 |
- |
- |
PREDICTED: uncharacterized protein LOC105645779 [Jatropha curcas] |
| 15 |
Hb_146673_010 |
0.1040264823 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 16 |
Hb_002184_110 |
0.1067389857 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 17 |
Hb_011930_020 |
0.1070351041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002820_050 |
0.1082282654 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 19 |
Hb_000059_380 |
0.1095901939 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 20 |
Hb_001623_270 |
0.1098169441 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |