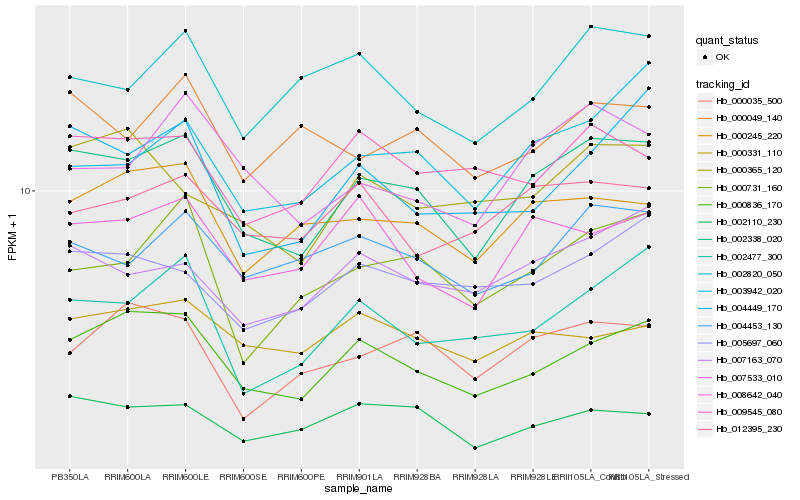
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002338_020 |
0.0 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 2 |
Hb_000836_170 |
0.0755021124 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 3 |
Hb_002110_230 |
0.0892593455 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 4 |
Hb_000245_220 |
0.0893441145 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 5 |
Hb_008642_040 |
0.0907233932 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007163_070 |
0.091385436 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_007533_010 |
0.0915786949 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 8 |
Hb_004453_130 |
0.0928391091 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000731_160 |
0.0986575817 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 10 |
Hb_005697_060 |
0.098660083 |
transcription factor |
TF Family: HB |
PREDICTED: pathogenesis-related homeodomain protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_004449_170 |
0.0996154084 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002820_050 |
0.0996317539 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 13 |
Hb_000331_110 |
0.0997371716 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 14 |
Hb_009545_080 |
0.1005148413 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 15 |
Hb_003942_020 |
0.1011769692 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000365_120 |
0.1017409379 |
- |
- |
PREDICTED: bifunctional protein FolD 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_012395_230 |
0.1018094328 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 18 |
Hb_002477_300 |
0.1020775063 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000049_140 |
0.1033128325 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000035_500 |
0.103337348 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 isoform X2 [Tarenaya hassleriana] |