| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
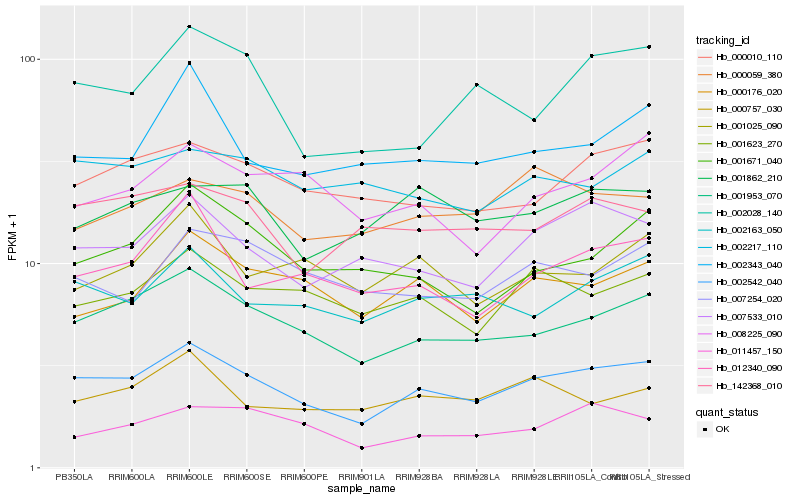
Hb_002542_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 2 |
Hb_001953_070 |
0.0707128269 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007533_010 |
0.0951125553 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 4 |
Hb_002163_050 |
0.0979841781 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |
| 5 |
Hb_000010_110 |
0.1002580797 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001623_270 |
0.104043505 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 7 |
Hb_012340_090 |
0.1051450937 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 8 |
Hb_000176_020 |
0.1081328818 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 9 |
Hb_002028_140 |
0.108701822 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 10 |
Hb_142368_010 |
0.110334084 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_002343_040 |
0.1120492181 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000757_030 |
0.1141332891 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 13 |
Hb_001862_210 |
0.1141933281 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 14 |
Hb_001671_040 |
0.11463821 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 15 |
Hb_011457_150 |
0.1147121765 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 16 |
Hb_007254_020 |
0.1148061695 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 17 |
Hb_000059_380 |
0.1152097082 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 18 |
Hb_002217_110 |
0.1162604695 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 19 |
Hb_001025_090 |
0.1165748413 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 20 |
Hb_008225_090 |
0.1176398203 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |