| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
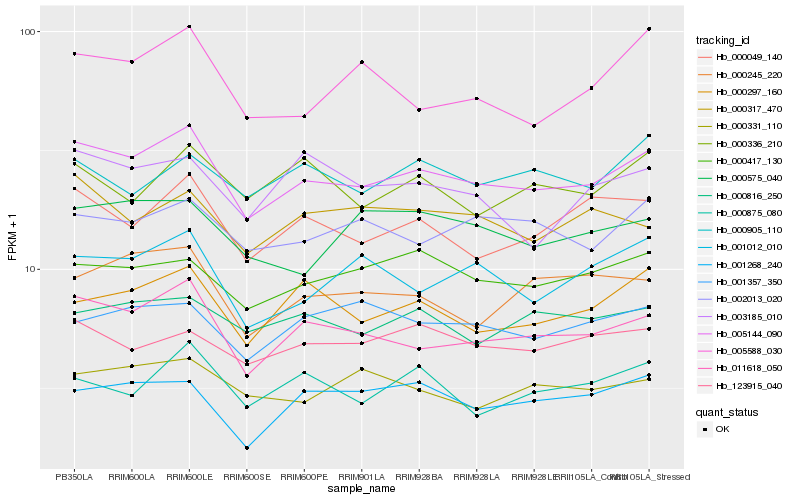
Hb_005144_090 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 2 |
Hb_011618_050 |
0.0475334975 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 3 |
Hb_000049_140 |
0.0664396303 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000417_130 |
0.0710378855 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_000297_160 |
0.0713533406 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_002013_020 |
0.0747724404 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 7 |
Hb_123915_040 |
0.0749870888 |
- |
- |
PREDICTED: uncharacterized protein LOC105633952 [Jatropha curcas] |
| 8 |
Hb_000816_250 |
0.0764939196 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001012_010 |
0.0775241841 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 10 |
Hb_000317_470 |
0.0785391284 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 11 |
Hb_000245_220 |
0.079030227 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 12 |
Hb_001268_240 |
0.0793112272 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001357_350 |
0.0804019694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000575_040 |
0.0804979238 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 15 |
Hb_000331_110 |
0.0810539102 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 16 |
Hb_000336_210 |
0.0820829967 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000875_080 |
0.082594456 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_003185_010 |
0.0828430263 |
- |
- |
PREDICTED: V-type proton ATPase subunit c''1 [Jatropha curcas] |
| 19 |
Hb_000905_110 |
0.0831380891 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 20 |
Hb_005588_030 |
0.0834761155 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |