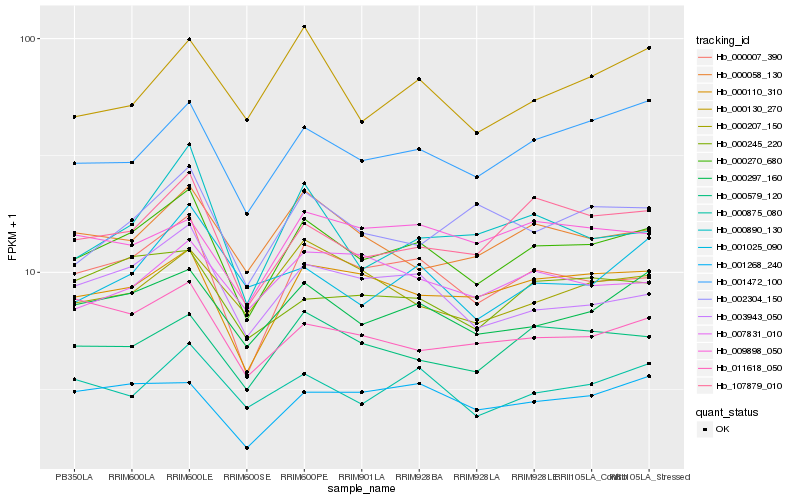
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_680 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 2 |
Hb_000007_390 |
0.0571440738 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 3 |
Hb_000297_160 |
0.0638435275 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_000110_310 |
0.0686443591 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_107879_010 |
0.0709629877 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 6 |
Hb_003943_050 |
0.0714733846 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 7 |
Hb_001472_100 |
0.0732512446 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 8 |
Hb_000579_120 |
0.0736583814 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 9 |
Hb_000875_080 |
0.0782415911 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_000245_220 |
0.0824309375 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 11 |
Hb_000058_130 |
0.0844821733 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 12 |
Hb_001268_240 |
0.0857097364 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000890_130 |
0.0885205514 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 14 |
Hb_011618_050 |
0.0901042025 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 15 |
Hb_000207_150 |
0.0921411928 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 16 |
Hb_001025_090 |
0.0924465366 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 17 |
Hb_002304_150 |
0.0927648543 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000130_270 |
0.0932999861 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_007831_010 |
0.0933550614 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 20 |
Hb_009898_050 |
0.0938902052 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |